Multiplexed polymerase chain reaction for genetic sequence analysis
A chain reaction and polymerase technology, which is applied in the field of multiplex polymerase chain reaction for genetic sequence analysis, can solve the problem of insensitivity to sequence diversity, difficulty in distinguishing, and inability to distinguish closely related strains of the same organism And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
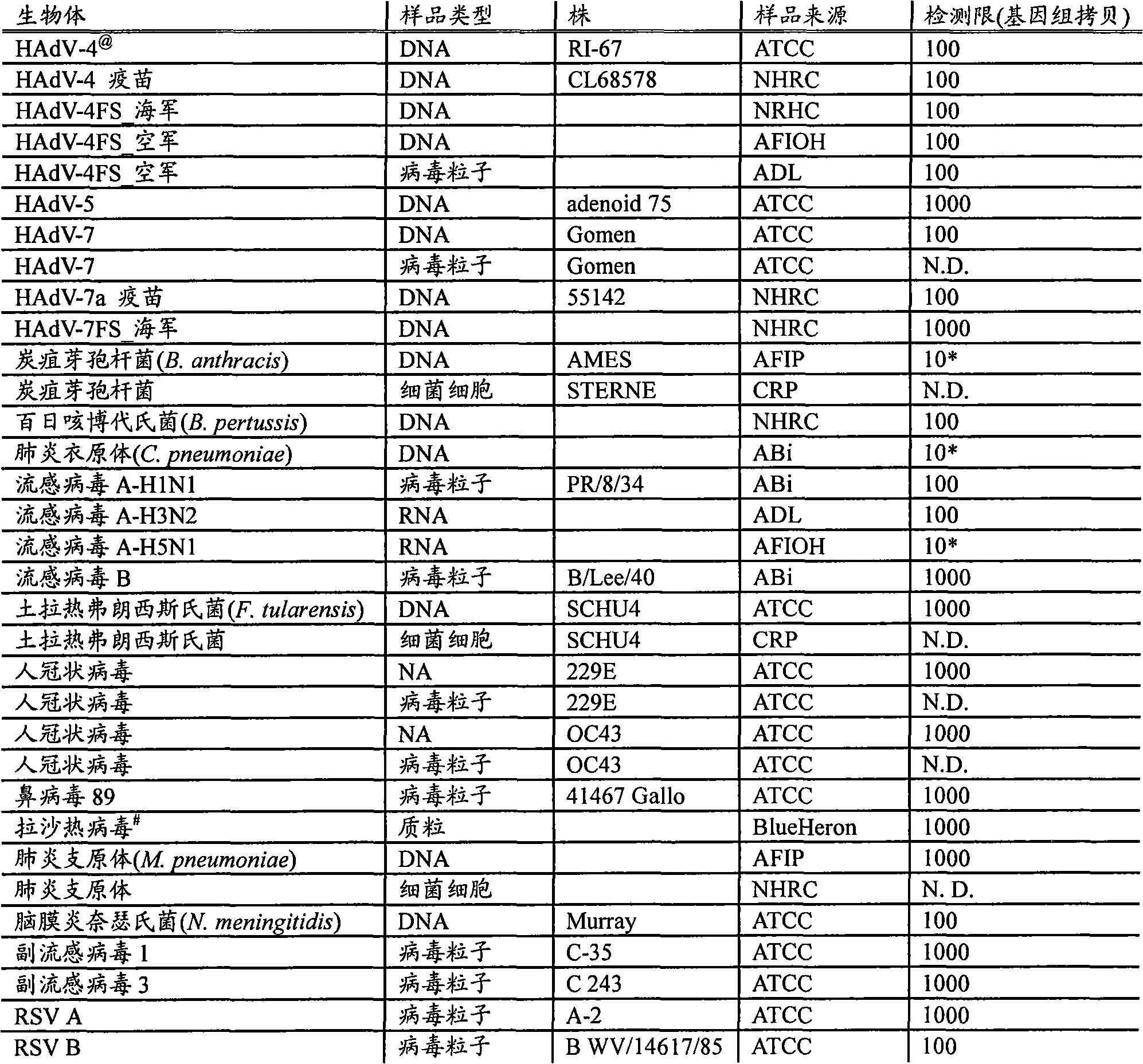
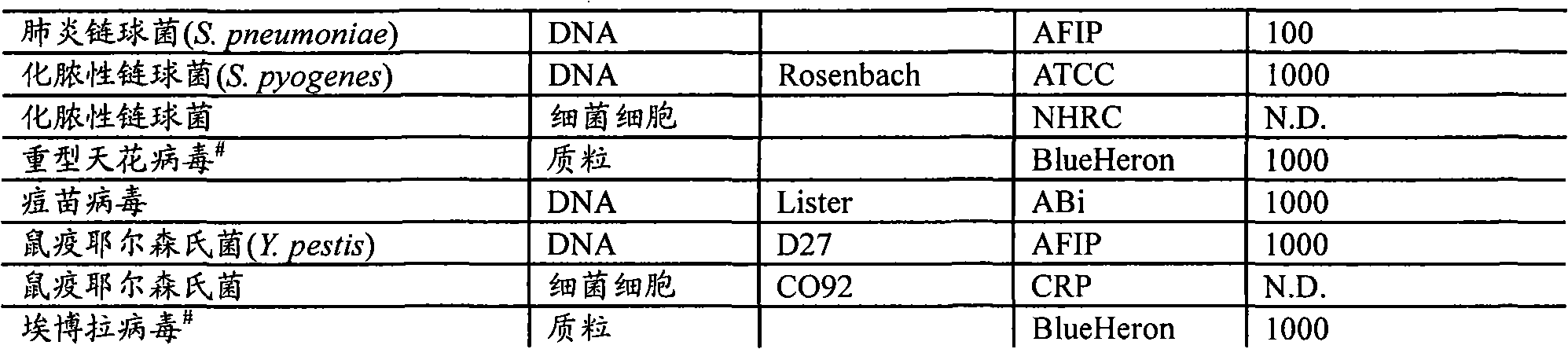
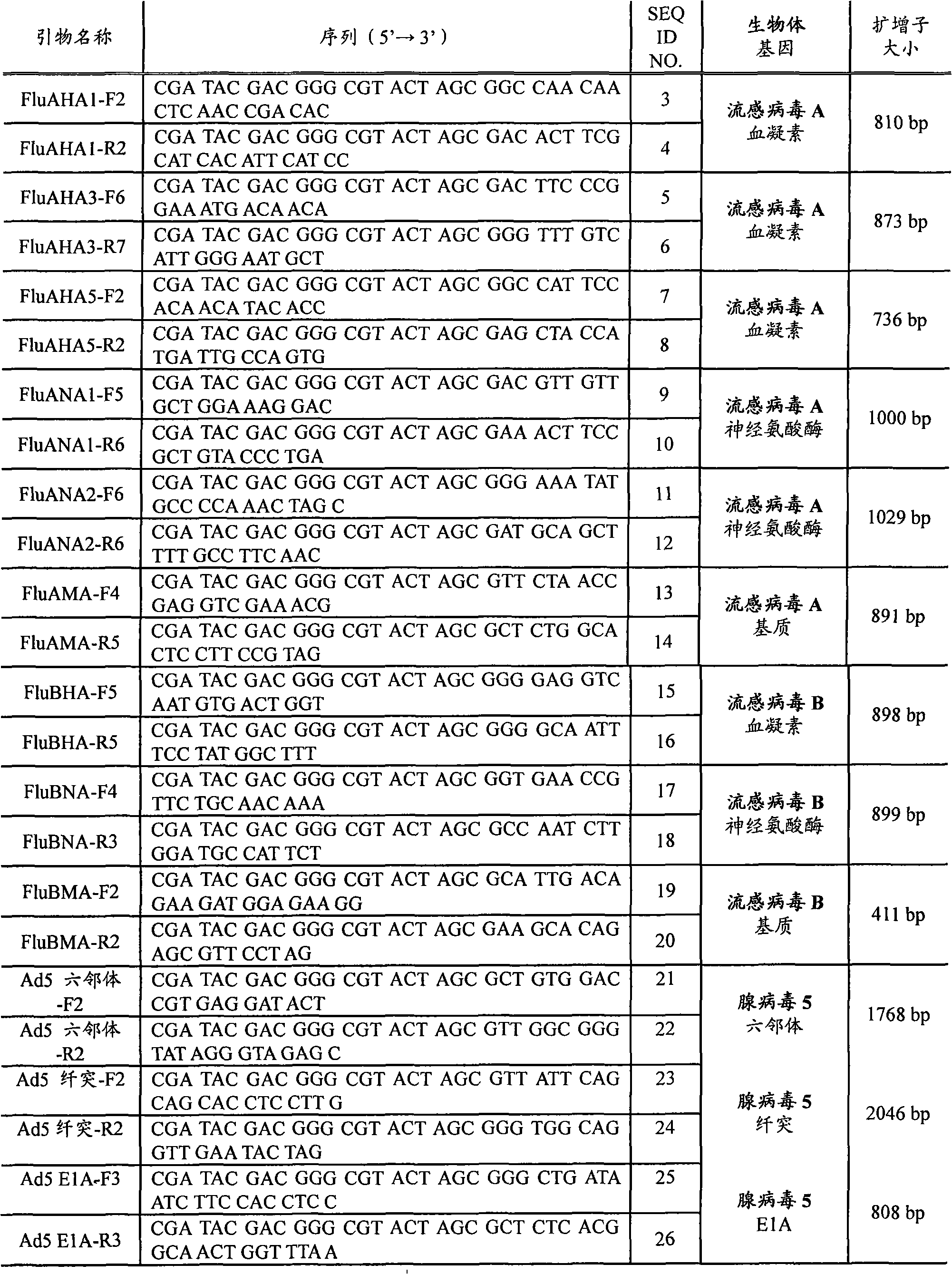
[0022] RPM v.1 Chip Design - The RPM v.1 (Respiratory Pathogen Microarray) chip design includes 57 tiles that allow resequencing of 29.7kb sequences from 27 respiratory pathogens and biological warfare agents and was detailed in previous studies Described (Lin et al., "Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays" Genome Res., 16(4), 527-535 (2006)). Briefly, RPM arrays consist of 25-mer perfectly matched probes representing sequences at each base within (and centered around) sequences selected from the genome of a target organism. In addition, for each perfectly matched probe, three mismatched probes representing the three possible single nucleotide polymorphisms (SNPs) at the central location were also tiled on the array. Thus, hybridization to a series of perfectly matched probes provides redundant presence / absence information, while hybridization to mismatched probes reveals strain-specific SNP data. On the chip, two pathogen...
Embodiment 2
[0028] Clinical samples-archived throat swabs were collected from patients with symptoms of ARI at multiple Army recruit training centers, the US-Mexico border zone, and on board Navy ships deployed in 1999-2005. These throat swabs were immediately placed in chilled 2 mL vials containing 1.5 mL of viral transport medium (VTM), frozen and stored at -80°C or below to maintain virions during transport. Samples were then shipped to the Naval Health Research Center (NHRC, San Diego, CA), thawed and aliquoted, and tested for HAdV and influenza virus using a CAP-certified diagnostic RT-PCR / PCR and culture assay. to test. Frozen aliquots were subsequently submitted for microarray-based detection in blinded fashion.
Embodiment 3
[0030] Nucleic Acid Extraction - Using MasterPure TM DNA purification kit (Epicentre Technologies, Madison, WI), omit RNA digestion, or use MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche Nucleic acids were extracted from clinical samples by Roche Applied Science, Indianapolis, IN, following the manufacturer's recommended protocol.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com