Method for detecting discrepancy expressed protein spectrum in blood serum sample of diabetes and kidney disease patient
A technology for diabetic nephropathy and differential expression, which is applied in the fields of biology and medical testing, and can solve problems such as limited application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
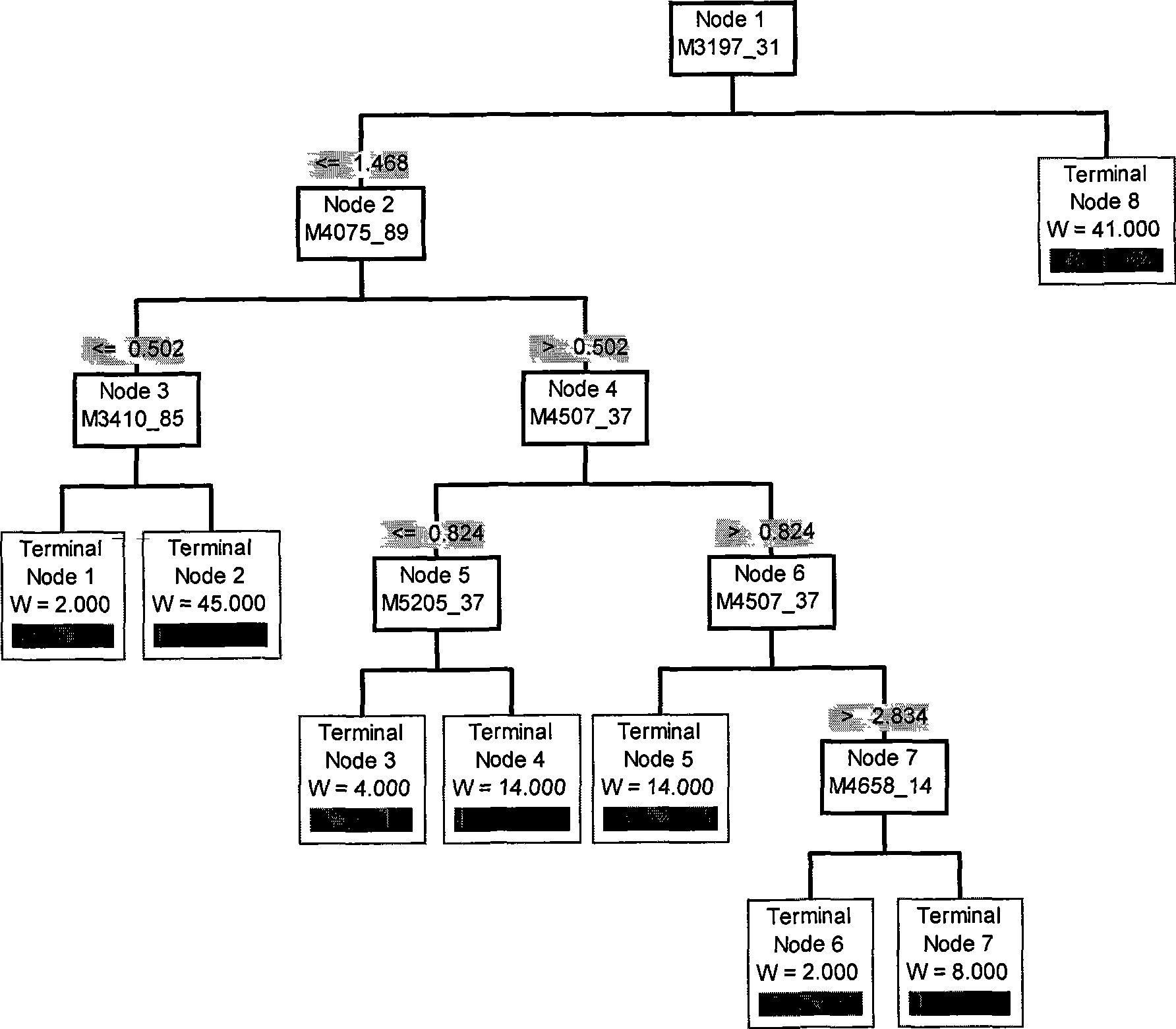
[0018] Example 1: Establishment of Serum Protein Fingerprint and DN Decision Tree Model of Type 2 Diabetic Nephropathy
[0019] Take serum samples from patients with type 2 diabetes and normal people:
[0020] Patients diagnosed with type 2 diabetic nephropathy, patients diagnosed with type 2 diabetes without nephropathy, normal people, fasting venous blood 5ml (without jaundice, lipemia, hemolysis, etc.), after 3 hours at room temperature, centrifuge at 3000rpm at 4°C for 10min, take The upper serum was stored in a -80°C refrigerator.
[0021] Quantitative protein concentration by Bradford method: blank control: 1ml Coomassie brilliant blue solution + 100μl deionized water; prepare standard curve according to Table 1; measure sample concentration; protein sample plus corresponding binding buffer (WCX buffer or SAX buffer or IMAC buffer solution or H4 buffer), diluted to 1mg / ml and loaded onto the corresponding chip.
[0022] Table 1 Preparation of standard curve
[0023] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| laser intensity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| laser intensity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com