Upland cotton disease-resistant gene and use thereof
A disease-resistant, transgenic plant technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problems of undeveloped upland cotton disease resistance gene npr1 and other problems, and achieve the effect of improving disease resistance and broad application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Cloning of Gh-npr1 gene
[0058] 1.2.1 Cloning of Gh-npr1 gene fragment
[0059]Cotton DNA was extracted by the improved CTAB method, and a degenerate primer p-F1(5'agg cac(t)tt(g)g ac(t)t cng atg ata(g)tt( a)g a3'), p-R1(5'tcc(t)c(t)tc(a,t)cg(t)ca ata(c)gcagca at(c)g(a)tga ag3'), Gene fragments were amplified by Touch-Down PCR. The PCR reaction conditions were: denaturation at 94°C for 5min; 2 cycles of 94°C for 30sec, 60°C for 30sec, and 72°C for 1min; 2 cycles of 94°C for 30sec, 58°C for 30sec, and 72°C for 1min; 94°C for 30sec, 56°C for 30sec, 72°C 1min, 2 cycles; 94°C 30sec, 54°C 30sec, 72°C 1min, 2 cycles; 94°C 30sec, 52°C 30sec, 72°C 1min, 30 cycles; 72°C extension 5min. 1.2.2 Obtaining the 3' end of cDNA of Gh-npr1 gene.
[0060] A specific primer F2 was designed according to the obtained gene fragment: 5'taactttggatgatgctactgcactccat3'. The leaf RNA of upland cotton 7124 was extracted by thermal CTAB method. Referring to the 3'RACE (TaKaRa, Japa...
Embodiment 2
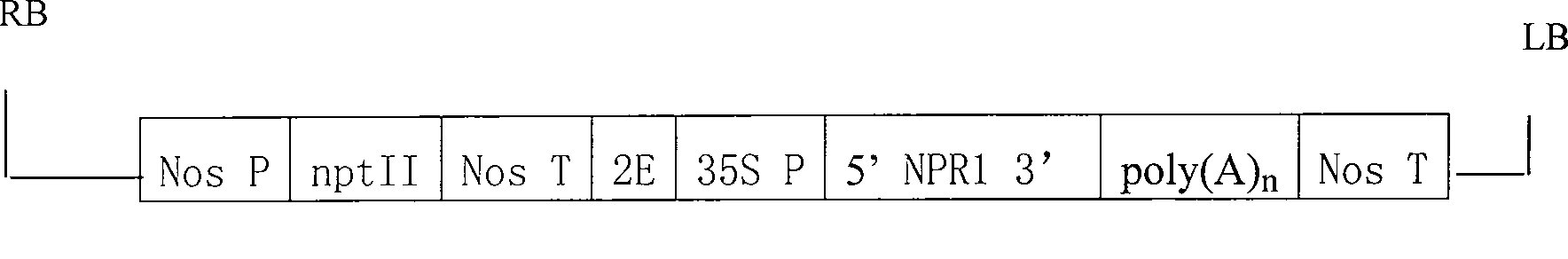
[0065] Example 2 Construction of Plant Expression Vectors
[0066] According to the obtained upland cotton npr1 cDNA sequence, primers p-F3 (5'ggcctcgagatggcttatttgtctgagccatcatct 3') and p-R4 (5'cgtctcgagtcacaatttcctatacttgtagg 3') were designed respectively. For the convenience of vector construction, Xho I restriction sites were introduced at the 5' end of p-F3 and the 3' end of p-R4. The first-strand cDNA obtained by reverse transcription of leaf RNA was used as a template for PCR amplification. The reaction conditions were: denaturation at 94°C for 4 min; 30 cycles of 45 s at 94°C, 45 s at 62°C, and 1.5 min at 72°C; extension at 72°C for 5 min. The target gene fragment was ligated into pMD18-T-Vector to obtain pMD-npr1, the pMD-npr1 plasmid was digested with Xho I, and the Gh-npr1 gene fragment was recovered for future use. Digest pBI121 and pTΩ4A with EcoRI / Hind III, recover 12kb and 1.4kb bands respectively, and construct the intermediate vector pB4A after ligation. T...
Embodiment 3
[0067] Example 3 Agrobacterium-mediated method to obtain bivalent transgenic tobacco
[0068] In this example, the transgenic tobacco with the Gh-npr1 gene was successfully obtained by using the Agrobacterium-mediated method.
[0069] The tobacco receptor material used is NC89, and the Agrobacterium is LBA4404. The specific operation steps are as follows:
[0070] 1) Preparation of competent Agrobacterium tumefaciens LBA4404
[0071] (1) Pick a single colony from the plate, inoculate it into 5ml YEB liquid medium (containing Streptomycin Strep 125mg / L), and culture overnight at 28°C and 250rpm with shaking;
[0072] (2) Take 2ml of bacterial liquid, add it into 50ml of YEB liquid medium (containing Strep125mg / L), shake and cultivate to OD at 28°C and 250rpm 600 About 0.6 or so;
[0073] (3) Transfer the bacterial solution to a 50ml sterile centrifuge tube and place in an ice bath for 30min. Centrifuge at 5000rpm for 5min;
[0074] (4) Discard the supernatant and use 2ml o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com