Induction of xa27 by the avrxa27 gene in rice confers broad-spectrum resistance to xanthomonas oryzae pv. oryzae and enhanced resistance to xanthomonas oryzae pv. oryzicola
A xa27, genome technology, applied in genetic engineering, botanical equipment and methods, biochemical equipment and methods, etc., can solve problems such as non-detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Materials and methods
[0070] Plasmids, bacterial strains and growth conditions
[0071] The plasmids used in this study were cosmid 99-avrXa27-20 (Gu et al., 2005) and pHM1avrXa27, cloning vector pGEM-T-easy (Promega, WI 53711, USA) and plant transformation vector pC1305.1 (CAMBIA, Canberra, Australia) and pCPR1avrXa27 (preparation described herein). The bacterial strains used in this study were Escherichia coli strain DH10b (Carlsbad, CA92008, USA), Agrobacterium tumefaciens strain AGL1 (Lazo et al., 1991), Xanthobacterium oryzae strain PXO99 A , AXO1947, AXO1947(pHM1avrXa27), K202, ZHE173 and CIAT1185, and B. oryzae strains L8(pHM1) and L8(pHM1avrXa27). Escherichia coli was cultured on Luria-Bertani medium at 37 °C; Agrobacterium tumefaciens strains were cultured on YEB medium at 28 °C; rice bacterial blight strains and rice bacterial spot bacterial strains were cultured on PSA medium at 28 °C (Gu et al. , 2004). The plasmids were introduced into E. coli, A. tum...
Embodiment 2
[0085] Construction of pCPR1avrXa27
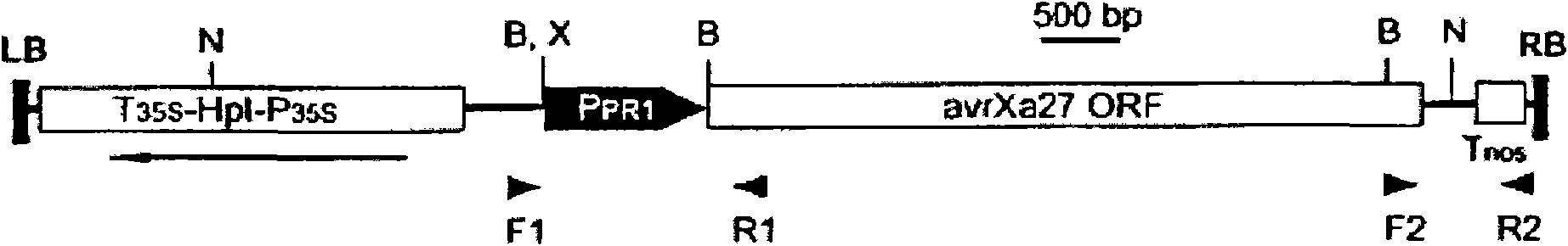
[0086] pCPR1avrXa27 was constructed based on CAMBIA vector pC1305.1. A 3482-bp genomic clone of the avrXa27 gene (including the 3411-bp full-length coding sequence (SEQ ID NO: 5)) from the cosmid 99-avrXa27-20 (Gu et al., 2005) was subcloned into pGEM-T-easy Vector (Promega), the intermediate construct pTavrXa27 was generated. The 3234-bp BamHI fragment of the avrXa27 gene in pTavrXa27 was replaced with the BamHI fragment of the avrXa27F2H gene from pZWavrXa27 (Gu et al., 2005), generating pTZWavrXa27. The SacII-AseI fragment from pTZWavrXa27 (containing the avrXa27 gene (SEQ ID NO: 1)) was isolated, blunt-ended, and cloned into the rice PR1 promoter in pC1305.1 (Gu et al., 2005; SEQ ID NO: 5 ) downstream, resulting in pCPR1avrXa27 ( figure 1 ). The primer pair used to determine the presence of the PR1 promoter and the nopaline synthase gene (Tnos) in transgenic plants were PF / PR (5'-CCATGATTACGAATTCGAGCTCGG-'3 (SEQ ID NO: 6) and 5'-AA...
Embodiment 3
[0088] Generation of avrXa27 transgenic plants
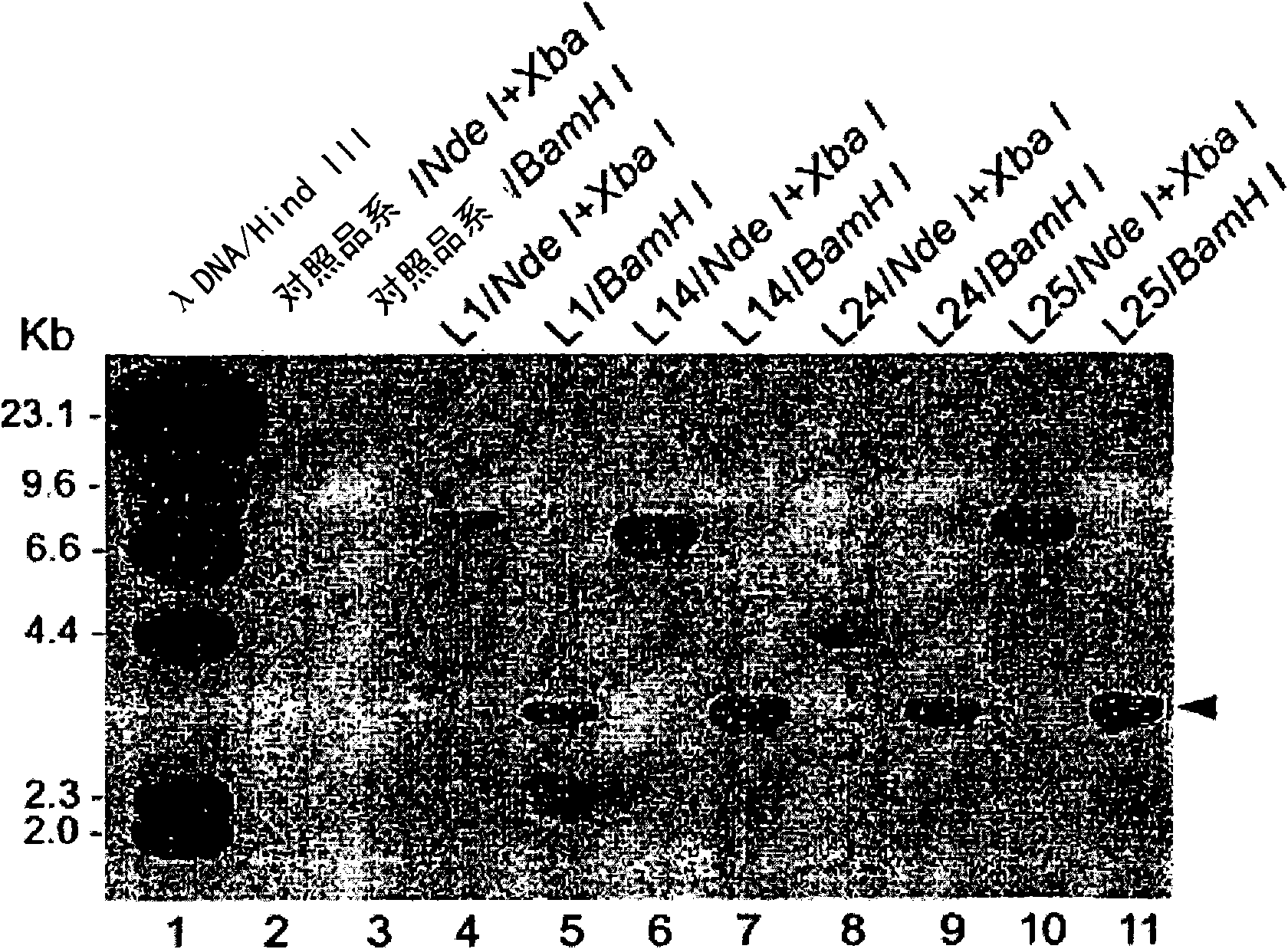
[0089] With the binary plasmid pCPR1avrXa27 ( figure 1 ) Agrobacterium-mediated transformation of the Nipponbare variety to generate avrXa27 transgenic plants. pCPR1avrXa27 carries P in its T-DNA region PR1 -αvrXα27-Tnos fusion gene. A total of 34 independent transgenic T 0 plants. However, after molecular analysis, only four strains of T 0 The plants (L1, L14, L24 and L25 lines) carried the complete αvrXα27 gene. Among the four transgenic plants ( figure 2 , lanes 5, 7, 9 and 11), Southern blot analysis detected the region corresponding to the coding region of αvrXα27 ( figure 1 3.2-kb band of predicted BαmHI fragment of ). At L24 ( figure 2 , lane 8), a 4.5-kb band corresponding to the predicted Nde I-XbαI fragment containing the PR1 promoter and avrXa27 coding region ( figure 1 ). However, this 4.5-kb NdeI-XbαI band was not detected in L1, L14, and L25. Instead, they all carry Nde I-XbαI bands with a molecular s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com