Method for accelerating RNA secondary structure prediction based on stochastic context-free grammar
A secondary structure and context technology, applied in the direction of machine execution devices, biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problem that does not involve three-dimensional dynamic programming matrix hardware acceleration, and has no RNA secondary structure prediction acceleration Methods and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
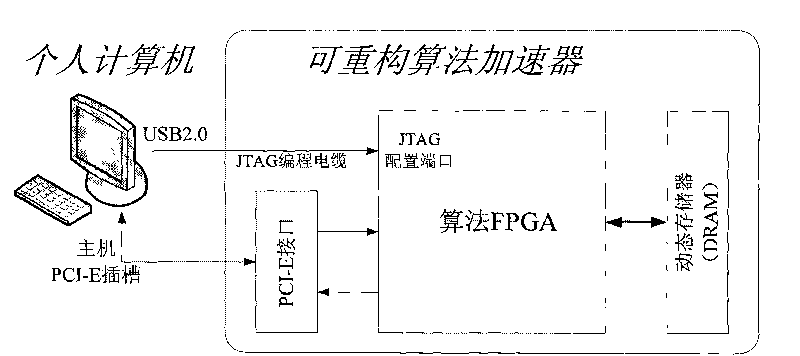
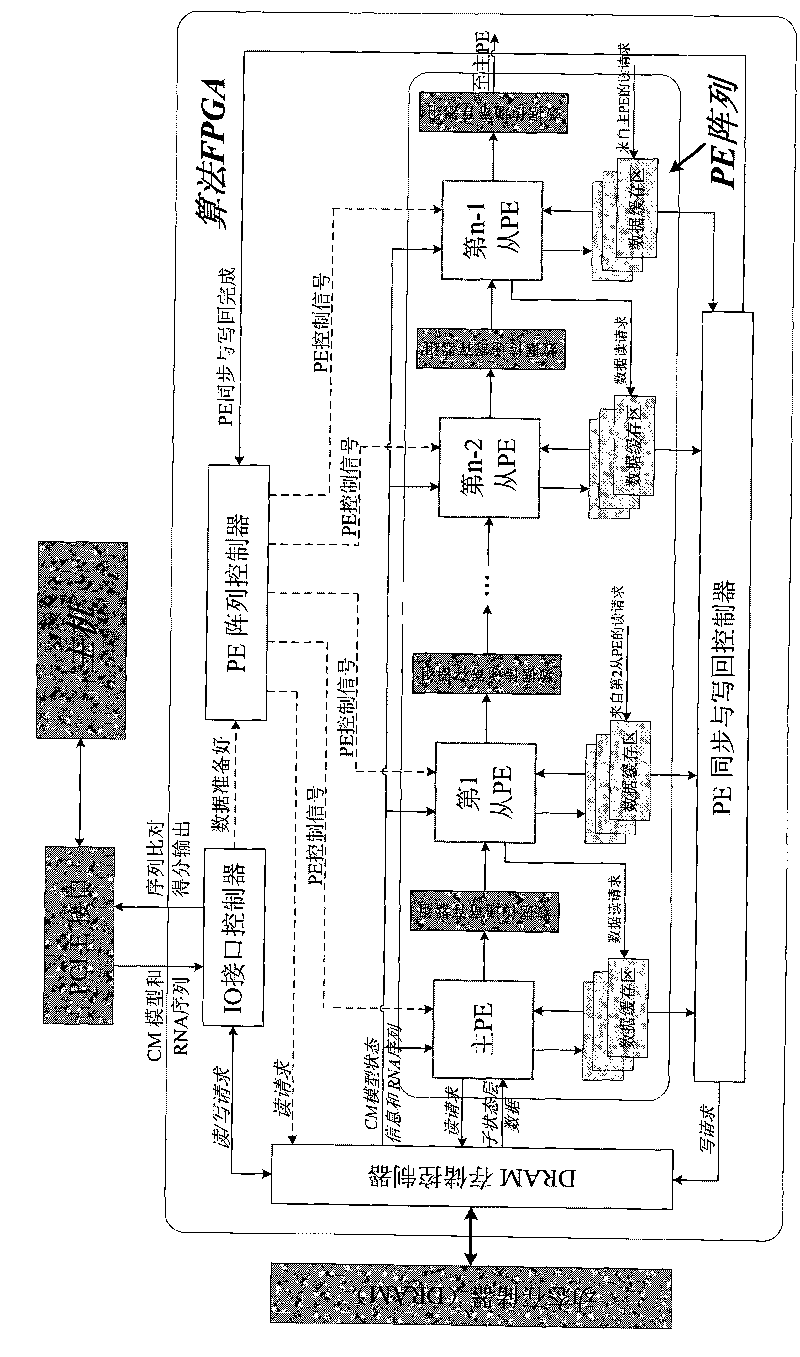
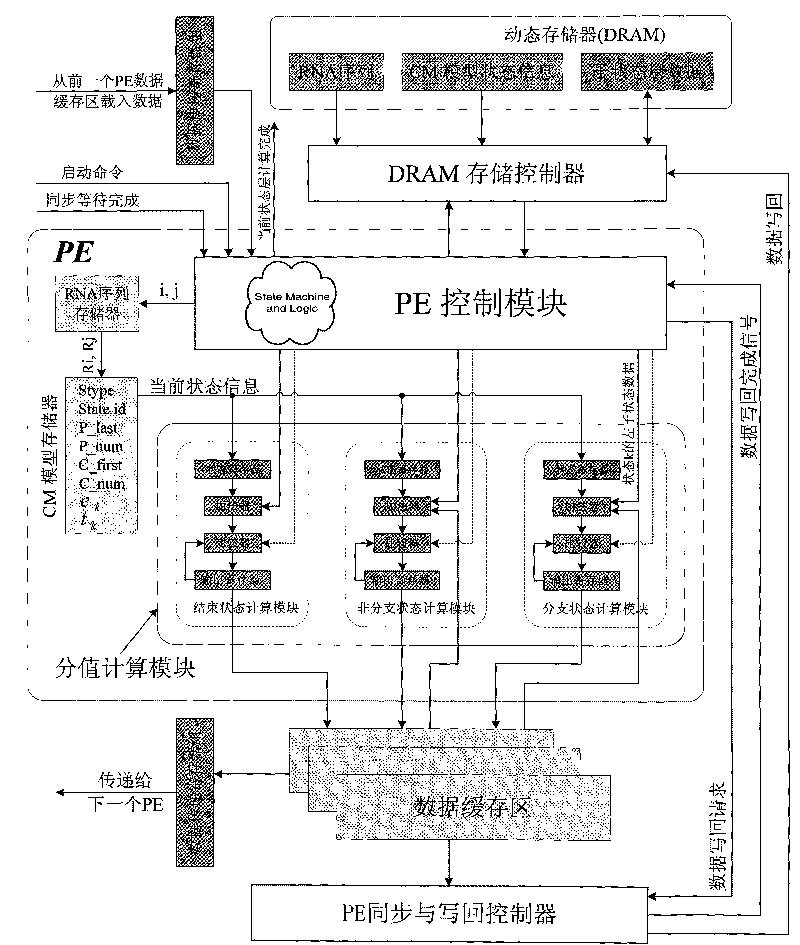
[0072] figure 1 It is the overall structure diagram of the heterogeneous computer system constructed in the first step of the present invention. The heterogeneous computer system consists of a host computer and a reconfigurable algorithm accelerator. The reconfigurable algorithm accelerator is inserted into the PCI-E slot of the host motherboard and connected to the host. A CM model formatting program, an RNA sequence coding program, a reconfigurable algorithm accelerator logic download program and a data communication program are installed on the host computer. The host configures the algorithm FPGA in the reconfigurable algorithm accelerator through the JTAG programming cable, loads the RNA sequence and CM model to the reconfigurable algorithm accelerator through the PCI-E interface, and starts the reconfigurable algorithm accelerator; the reconfigurable algorithm accelerator Complete the calculation of the CYK / inside algorithm without backtracking, and then return the co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com