Method for identifying cymbidium varieties
A variety and sequence technology, applied in the field of plant variety identification, can solve the problems of poor specificity and small number of varieties, and achieve the effect of short time, fast speed and DNA fingerprint identification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
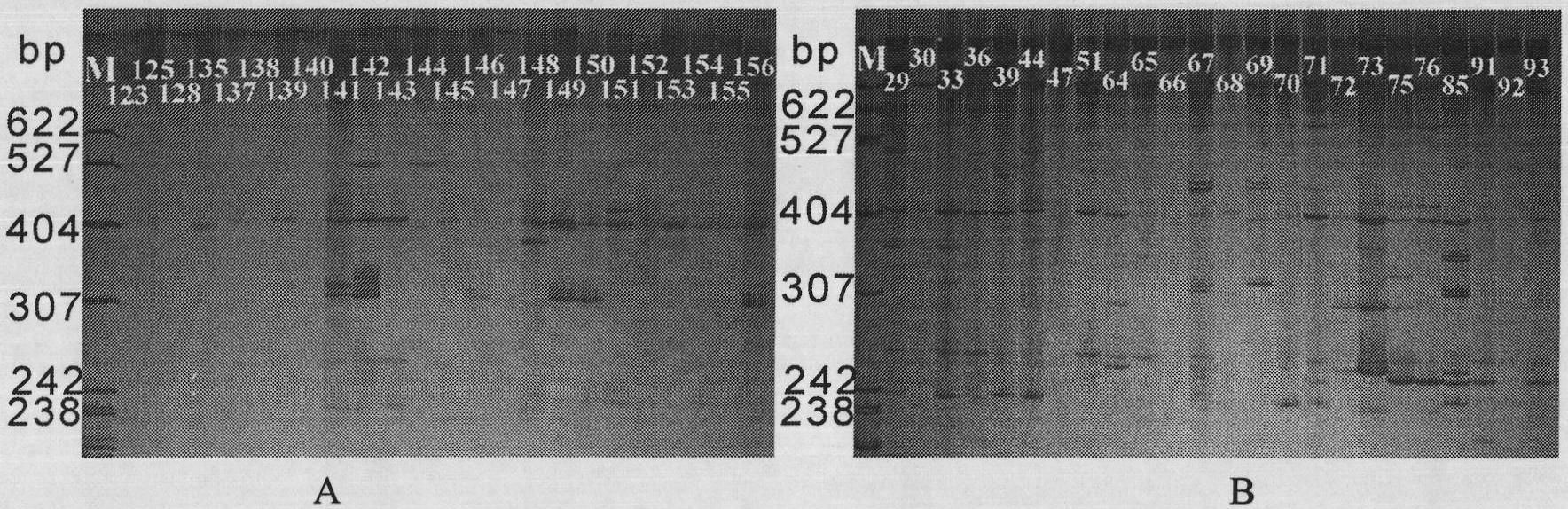
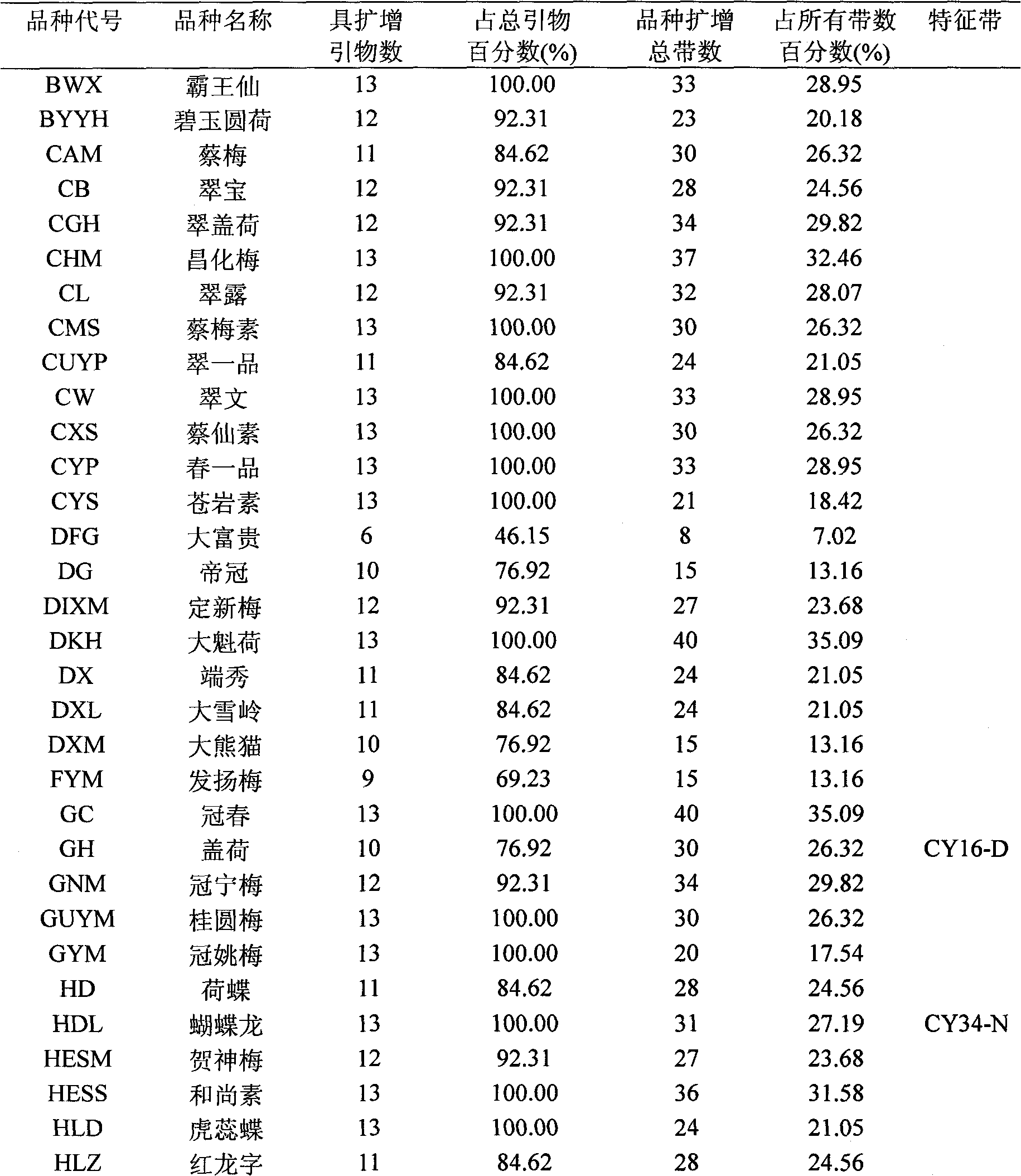
[0022] (1) Extract the genomic DNA of leaves of existing 112 Chunlan varieties; (2) carry out polymerase chain reaction (PCR reaction) respectively with the extracted Chunlan varieties genomic DNA to obtain simple repeat sequence products; (3) use 6% denaturation The polyacrylamide gel was used for electrophoresis separation of simple repetitive sequence products. The total loading volume was 5 μL, of which 4 μL was the amplification product, plus 1 μL Loading Buffer, and electrophoresed at a constant power of 60 W until the marker ran to 2 / 3 of the gel plate; After electrophoresis, take pictures with a digital camera by the Bassam silver staining method, and obtain the standard spectrum of simple repeat sequences respectively; (4) select a clear simple repeat sequence spectrum, use the "0-1" system to record the bands, and assign the bands to 1, the assignment of no band is 0; (5) read the data of the electrophoretic bands of the products amplified by different primers for eac...
Embodiment 2
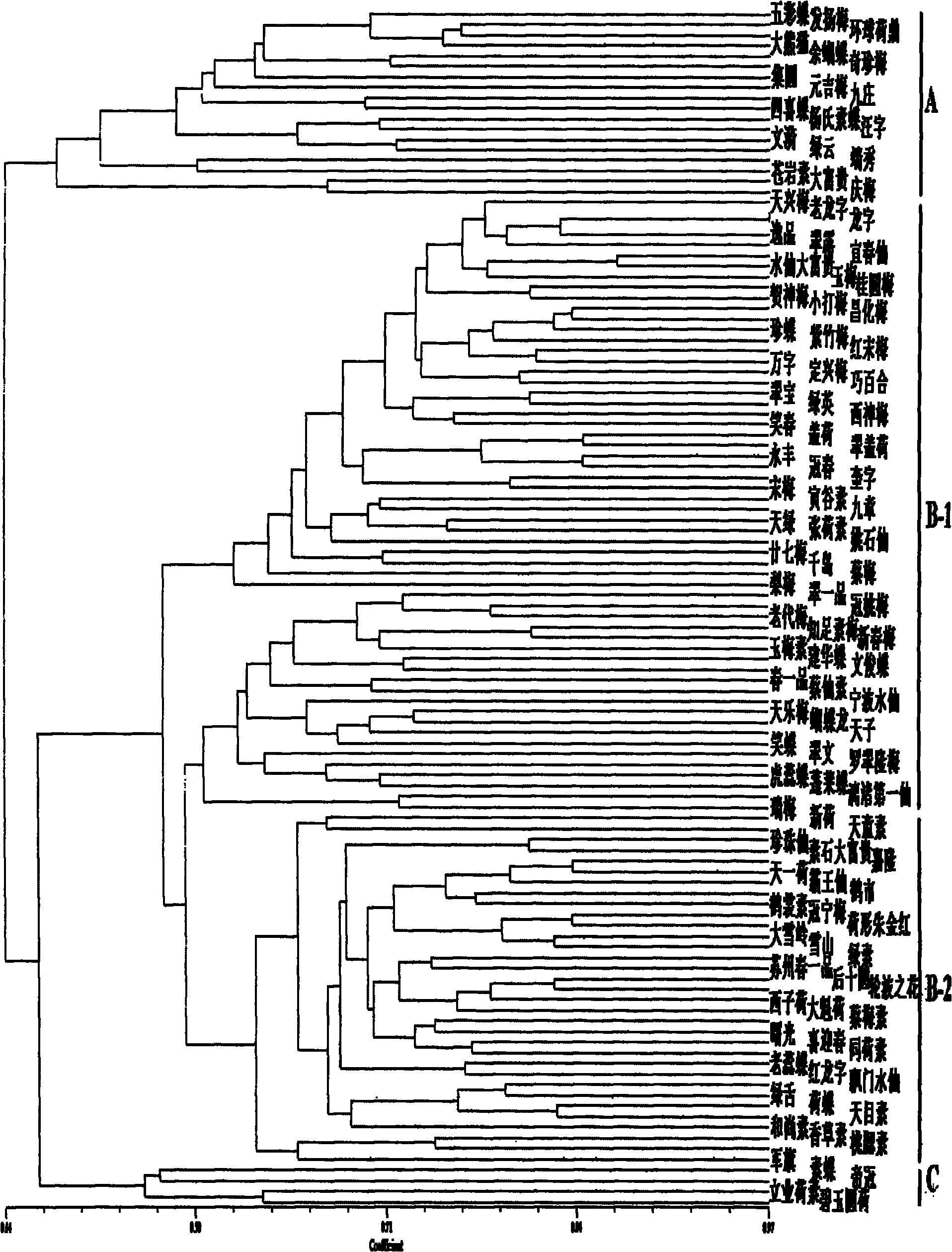
[0031] The 'Laolongzi' variety ( figure 2 The Chinese code is LLZ) for identification, through the combination of 13 pairs of primers, combined with the calculation of the similarity coefficient, it was included in the standard map database for cluster analysis, and it was found that it was similar to the 'Longzi' variety ( figure 2The Chinese code is LZ) tightly combined. Statistical tests were performed on the fingerprints of LZ and LLZ, and the P value of the paired chi-square test of the contingency table was 1, indicating that the two varieties had the same fingerprints. The P value of the Kappa test is 0, so the null hypothesis is rejected and the two varieties are considered to be consistent. Combined with the Kappa index of 0.958, it can be considered that there is a very high consistency between them. Finally, it can be judged as the 'Dragon Character' variety. In conclusion, using one or more effective primers can distinguish and identify all Chunlan varieties in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com