Fluorescence complementary system based on green fluorescent protein sfGFP
A fluorescent and protein technology, applied in the field of fluorescent complementary systems, can solve problems such as easy self-activation and false positives, and achieve the effects of low false positive rate, good effect and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1, inventive concept
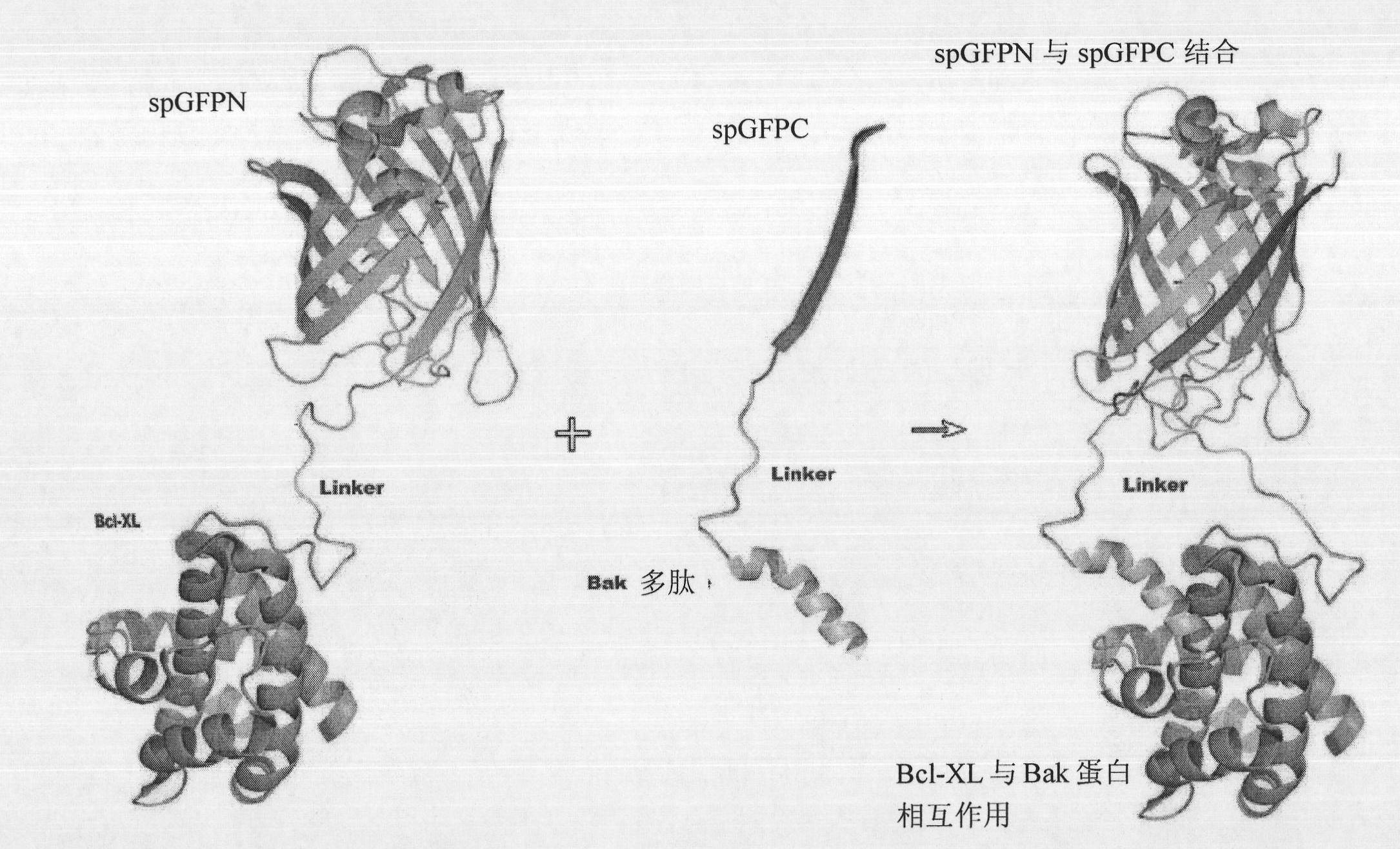
[0034] (1) if figure 1 As shown, the green fluorescent protein super fold GFP was split into GFP1-10βstrands (sfGFPN) and GFP11βstrands (sfGFPC) from 214 / 215 amino acid residues; Xl), fusion of protein B (such as Bak+) to be detected; if protein A and protein B interact, the separated sfGFPN and sfGFPC will reassemble into GFP that can emit fluorescence; if protein A and protein B do not interact, then Separated sfGFPN and sfGFPC do not reassemble and cannot fluoresce. (2) The present invention further performs point mutation on the sfGFPC fragment, and then detects the interaction between protein A and protein B according to the method described in (1). (3) From sfGFPC non-mutated and sfGFPC fragments with different point mutations, select sfGFPC fragments with strong fluorescence effect and low false positive to apply to the interaction detection of other proteins to be tested, thus completing the present invention.
[0035] It can...
Embodiment 2
[0043] Embodiment 2, the composition of the fluorescent complementary system constructed by the present invention
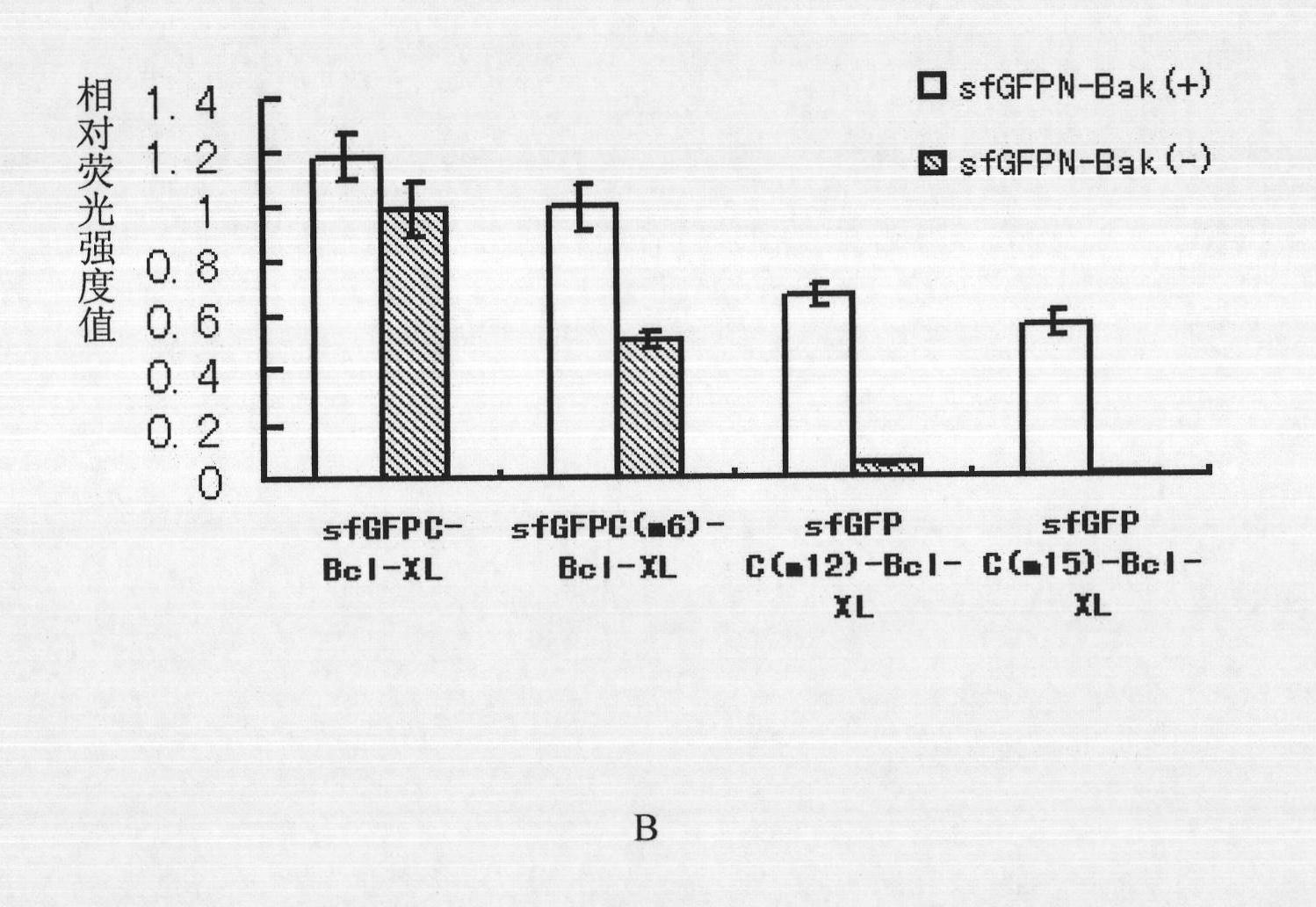
[0044] Each group of fluorescent complementary systems constructed by the present invention is composed of protein fragment I and protein fragment II, specifically as follows:
[0045] 1. Protein fragment I: shown in SEQUENCE ID: 1 (ie sfGFPN); protein fragment II: shown in SEQUENCE ID: 3 (ie sfGFPC);
[0046] 2. Protein fragment I: shown in SEQUENCE ID: 1 (ie sfGFPN); protein fragment II: shown in SEQUENCE ID: 5 (namely sfGFPC with point mutation, denoted as sfGFPC(m6));
[0047] 3. Protein fragment I: shown in SEQUENCE ID: 1 (ie sfGFPN); protein fragment II: shown in SEQUENCE ID: 7 (namely sfGFPC with point mutation, denoted as sfGFPC(m12));
[0048] 4. Protein fragment I: shown in SEQUENCE ID: 1 (ie sfGFPN); protein fragment II: shown in SEQUENCE ID: 9 (namely sfGFPC with point mutation, denoted as sfGFPC(m15));
Embodiment 3
[0049] Embodiment 3, the effect verification of each group of fluorescence complementary systems of the present invention
[0050] The primers used in the following experiments are shown in Table 1 and Table 2.
[0051] Plasmid pcDNA3.1 was purchased from Invitrogen, catalog number V860-20.
[0052] 1. Preparation of recombinant expression vectors expressing various fusion proteins
[0053] (1) Preparation of recombinant expression vector expressing fusion protein sfGFPC-Bcl-xL
[0054] 1. Preparation of sfGFPC gene
[0055] The sfGFPC gene was synthesized by annealing with primers Fold03c, Fold04n, Fold-05c and Fold-06n, and the two ends of the synthetic gene had NotI and ClaI restriction sites.
[0056] The basic process of annealing synthesis:
[0057] (1) Fragment treatment: primer concentration 0.1nmol / ul, Fold03c 10ul, Fold04n 10ul, Fold05c10ul, Fold06n 10ul, Ligase buffer (10×) 5ul, H 2 O 5ul, a total of 50ul. Boil for 5 minutes, cool to room temperature
[0058]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com