Method for identifying life or death of cells of food-borne pathogenic bacteria
A technology for food-borne pathogenic bacteria and pathogenic bacteria, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and the determination/inspection of microorganisms. Dead cells, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
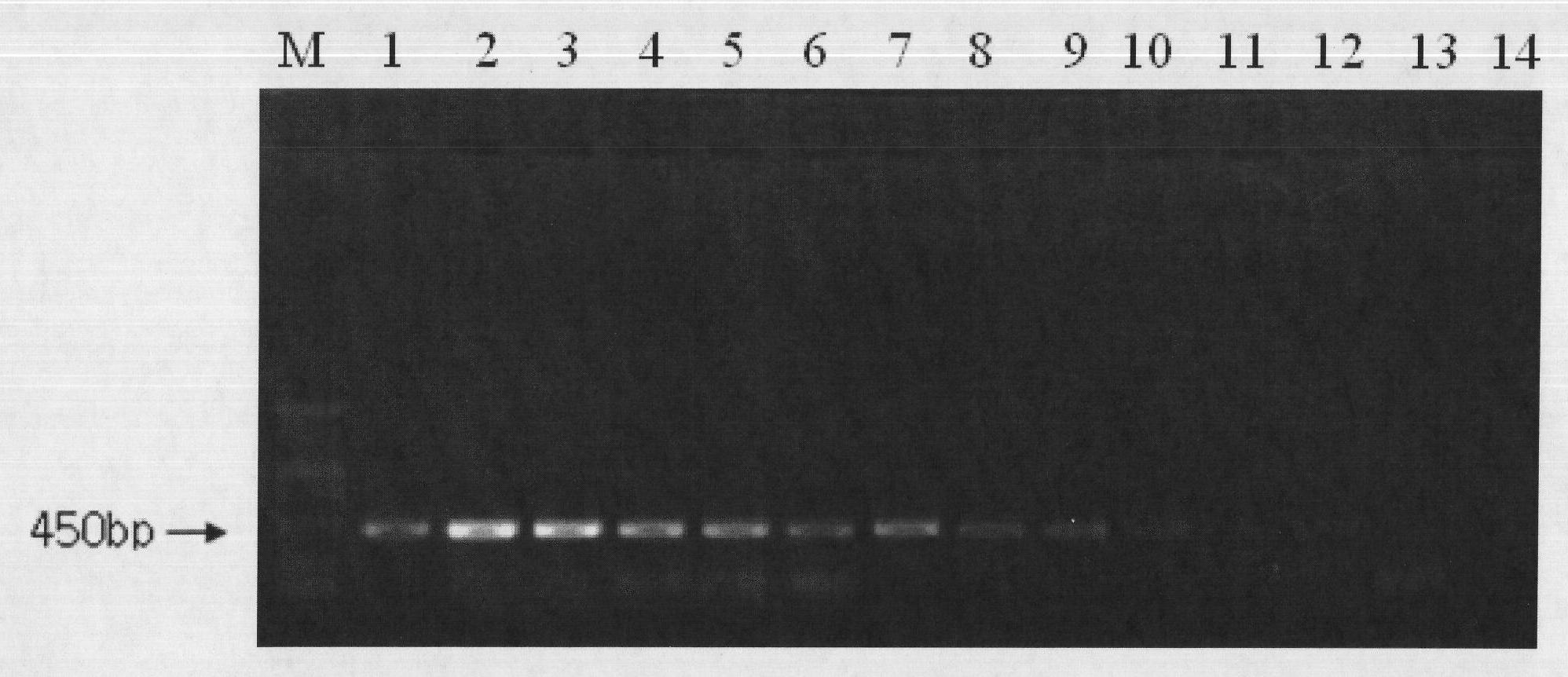
[0031] (1) EMA treatment of live / dead cells of Vibrio parahaemolyticus at different concentrations
[0032] Take 1 mL of Vibrio parahaemolyticus live / dead cell suspensions at different concentrations and place them in sterile centrifuge tubes, add EMA at a final concentration of 5-50 μg / mL and place them in the dark for 3-10 min, place the centrifuge tubes in In an ice bath with the tube cap opened, expose to a 650W halogen lamp for 5-15 minutes.
[0033] (2) Preparation of DNA template of Vibrio parahaemolyticus after EMA treatment
[0034] Take 1 mL of the EMA-treated bacterial solution in a sterile centrifuge tube, centrifuge at 10,000 r / min for 5 min, discard the supernatant, wash 2 to 3 times with normal saline, add 100 μL of TE to resuspend, bathe in 100°C water for 10 min, and cool in an ice bath for 10 min. Centrifuge at 12000r / min for 5min, and take the supernatant as a PCR template.
[0035] (3) PCR amplification reaction
[0036] Primer synthesis: The primer sequ...
Embodiment 2
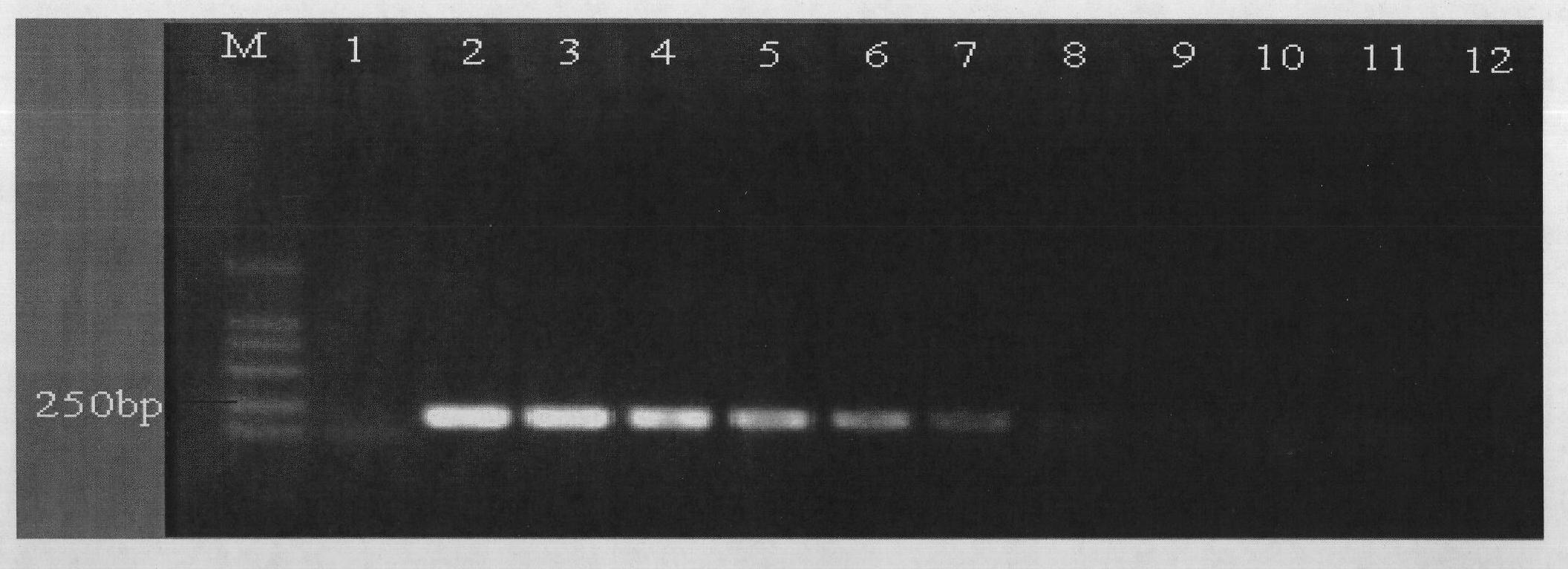
[0043] (1) EMA treatment of mixed bacterial suspensions of Vibrio parahaemolyticus with different proportions of viable cells
[0044] Vibrio parahaemolyticus live cell suspension and dead cell suspension (2.0×10 7 CFU / mL) to prepare viable cells containing 100%, 75%, 50%, 25%, 20%, 10%, 5%, 4%, 3%, 2%, 1%, 0% (both volume percentages) To the bacterial suspension mixed system, add EMA with a final concentration of 5-50 μg / mL and place in the dark for 3-10 minutes. Place the centrifuge tube in an ice bath and open the cap, and expose to a 650W halogen lamp for 5-15 minutes.
[0045] (2) Preparation of DNA template of Vibrio parahaemolyticus after EMA treatment
[0046] Take 1 mL of the EMA-treated bacterial solution in a sterile centrifuge tube, centrifuge at 10,000 r / min for 5 min, discard the supernatant, wash 2 to 3 times with normal saline, add 100 μL of TE to resuspend, bathe in 100°C water for 10 min, and cool in an ice bath for 10 min. Centrifuge at 12000r / min for 5min...
Embodiment 3
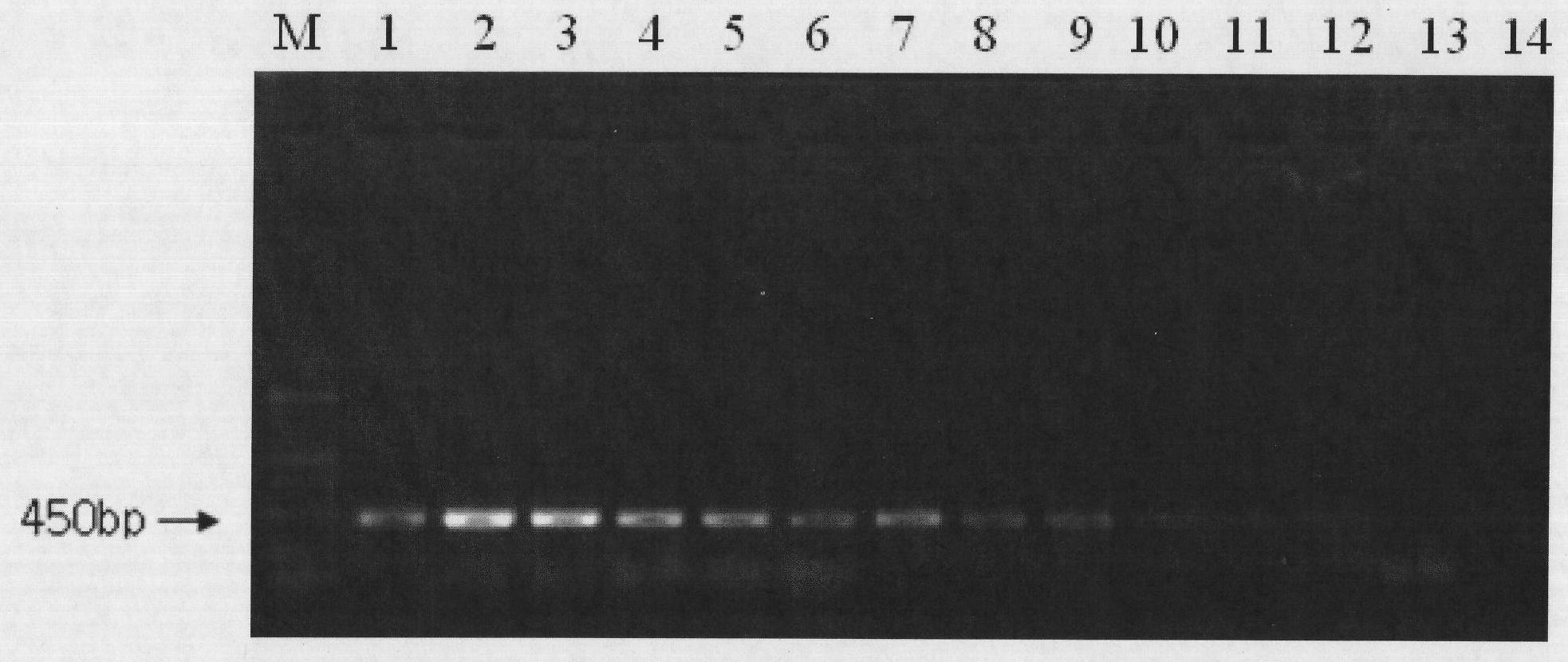
[0053] (1) EMA treatment of different concentrations of Shigella live / dead cells
[0054] Take 1 mL of Shigella live / dead cell suspensions at different concentrations and place them in sterile centrifuge tubes, add EMA at a final concentration of 5-50 μg / mL and place them in the dark for 3-10 min, place the centrifuge tubes in In an ice bath with the tube cap opened, expose to a 650W halogen lamp for 5-15 minutes.
[0055] (2) Preparation of Shigella DNA template after EMA treatment
[0056] Take 1 mL of the EMA-treated bacterial solution in a sterile centrifuge tube, centrifuge at 10,000 r / min for 5 min, discard the supernatant, wash 2 to 3 times with normal saline, add 100 μL of TE to resuspend, bathe in 100°C water for 10 min, and cool in an ice bath for 10 min. Centrifuge at 12000r / min for 5min, and take the supernatant as a PCR template.
[0057] (3) PCR amplification reaction
[0058] Primer synthesis: The primer sequences are as follows, and the length of the amplifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com