Mycoplasma contamination detection method and application
A mycoplasma and to-be-detected technology, applied in the biological field, can solve the problems of high price of mycoplasma detection kits, affecting the accuracy of results, and high use costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] The 23 kinds of mycoplasma obtained from the above screening and plasmid synthesis were selected as detection targets,
[0108] The sequences of the three upstream primers are: 5'-ACCACCCGGTAAGGCTAAATACTAACCAGAC-3' (Seq ID No.1) can amplify M.bovis, M.bovoculi, M.fermentans, M.synoviae, M.pulmonis, M.flocculare, M. hyorhinis, M.hominis, M.arginini, M.arthritidis, M.hyosynoviae, M.orale, M.salivarium, M.hyopneumoniae, Spiroplasma citri a total of 15 kinds of mycoplasma; 5'-AGGACCATCTCCTAAGGCTAAATACTACTTGG-3'(Seq ID No.2 ) can amplify Acholeplamasalaidlaawii; 5'-CAAACCATTGGGTAAGCCTAAATACTAGTCAG-3' (Seq ID No.3) can amplify M.gallisepticum, M.pirum, M.pneumoniae, M.penetrans, M.genitalium, Ureaplasmaurealyticum, M.capricolum A total of 7 kinds of mycoplasma.
[0109] The sequences of the two downstream primers are: 5'-CACCCATTAACGGGCTCTGACTAATTGTAA-3' (Seq ID No.4) can amplify M.bovis, M.bovoculi, M.fermentans, M.synoviae, M.pulmonis, M.flocculare, M. .hyorhinis, M.homin...
Embodiment 2
[0133] Select the primers and probe sequences designed in Example 1, carry out the amplification test of the method version of the recombinase polymerase amplification (in conjunction with endonuclease IV), and construct the amplification reaction system as follows:
[0134] 30mM tris-acetic acid buffer pH8.0
[0135] 100mM potassium acetate
[0136] 14mM magnesium acetate
[0137] 3mM Dithiothreitol
[0138] 5% polyethylene glycol (20000)
[0139] 3mM ATP
[0140] 30mM creatine phosphate
[0141] 100ng / ul creatine kinase
[0142] 400ng / ul E. coli recA protein
[0143] 200ng / ul E. coli SSB protein
[0144] 60ng / ul E. coli recO protein
[0145] 40ng / ul E. coli recR protein
[0146] 60ng / ul E. coli recF protein
[0147] 8Units Bacillus subtilis DNA polymerase I
[0148] 50ng / ul Endonuclease IV
[0149] 450uM dNTPs
[0150] 250uM each upstream primer
[0151] 250uM each downstream primer
[0152] 120nM fluorescent probe
[0153] The amplification temperature was 4...
Embodiment 3
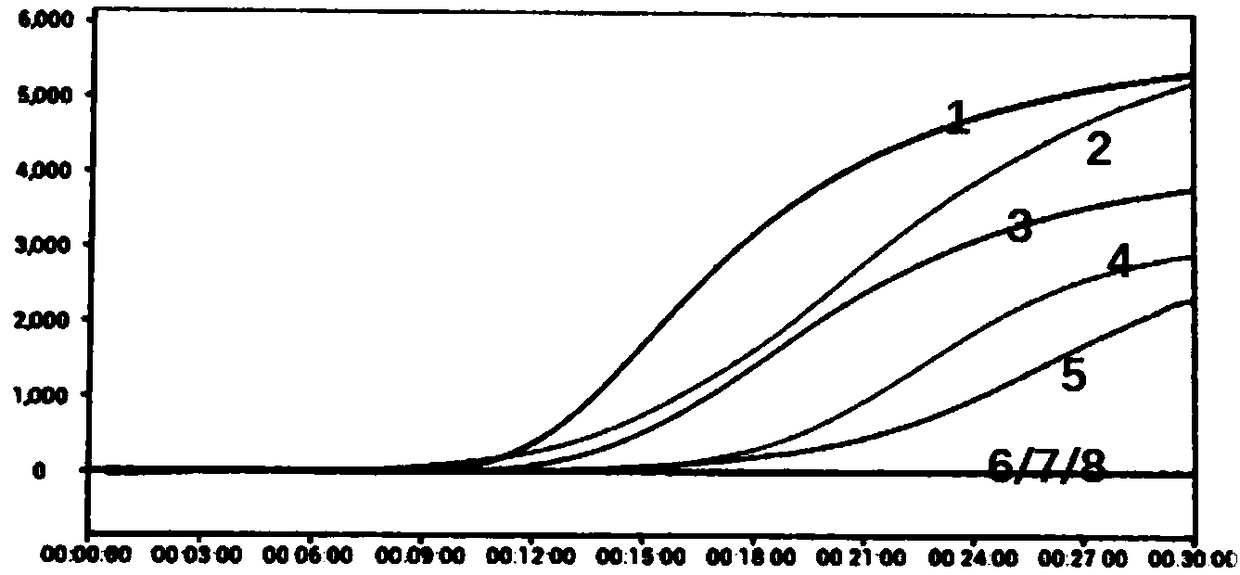
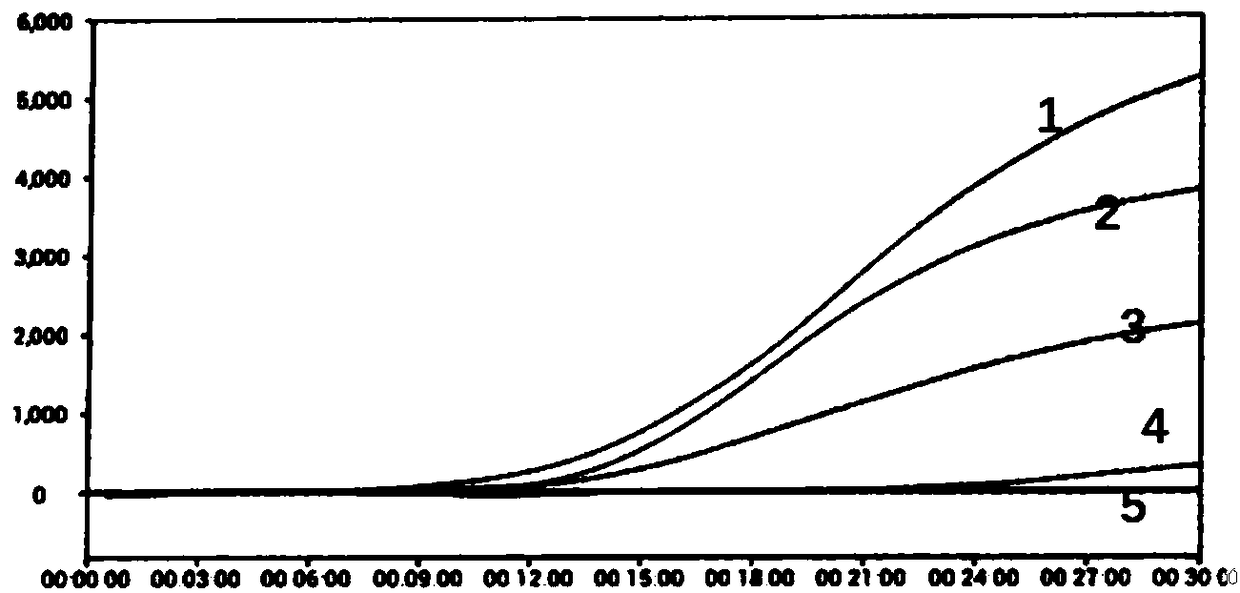
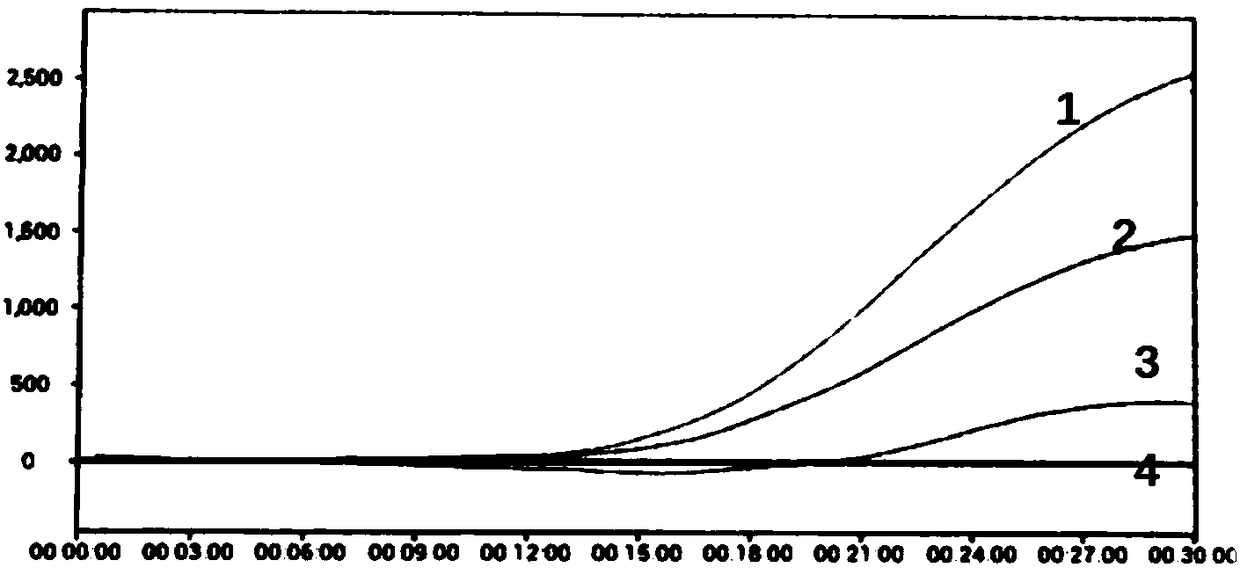
[0156] The design of all primers and probes gives priority to 8 common mycoplasmas (M.arginini, M.fermentans, M.hominis, M.orale, M.hyorhinis, M.salivarium, M.pirum and Acholeplamasalaidlaawii), and further Consider covering other mycoplasmas, and at the same time exclude the interference of E. coli, see the comparison results Figure 10 :
[0157] The probe and primer sequences are as follows:
[0158] Upstream primer sequence: 5'-CACCTCGATGTCGRCTCATCTCATCCTCGA-3' (Seq ID No 6), which can amplify all 23 kinds of mycoplasma;
[0159] The sequences of the three downstream primers are: 5'-CTAGGAACAGCTCTTCTCAATCTTCCTACGGG-3' (Seq ID No 7), which can amplify 6 species of mycoplasma including M.gallisepticum, M.pirum, M.pneumoniae, M.penetrans, M.capricolum, and Ureaplasma urealyticum ; 5'-GTAGCTCTCATCAATATTCCAACGCCCACAT-3'(SeqID No 8), can amplify M.bovis, M.bovoculi, M.fermentans, M.synoviae, M.pulmonis, M.flocculare, M.hyorhinis, M.hominis, M.arginini, M.arthritidis, M.hyosyn...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com