Method and system for protein purification
A protein-binding and recombinant protein technology, applied in the field of protein purification, can solve the problems of inconvenient protein purification process, enzyme loss, enzyme activity destruction, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
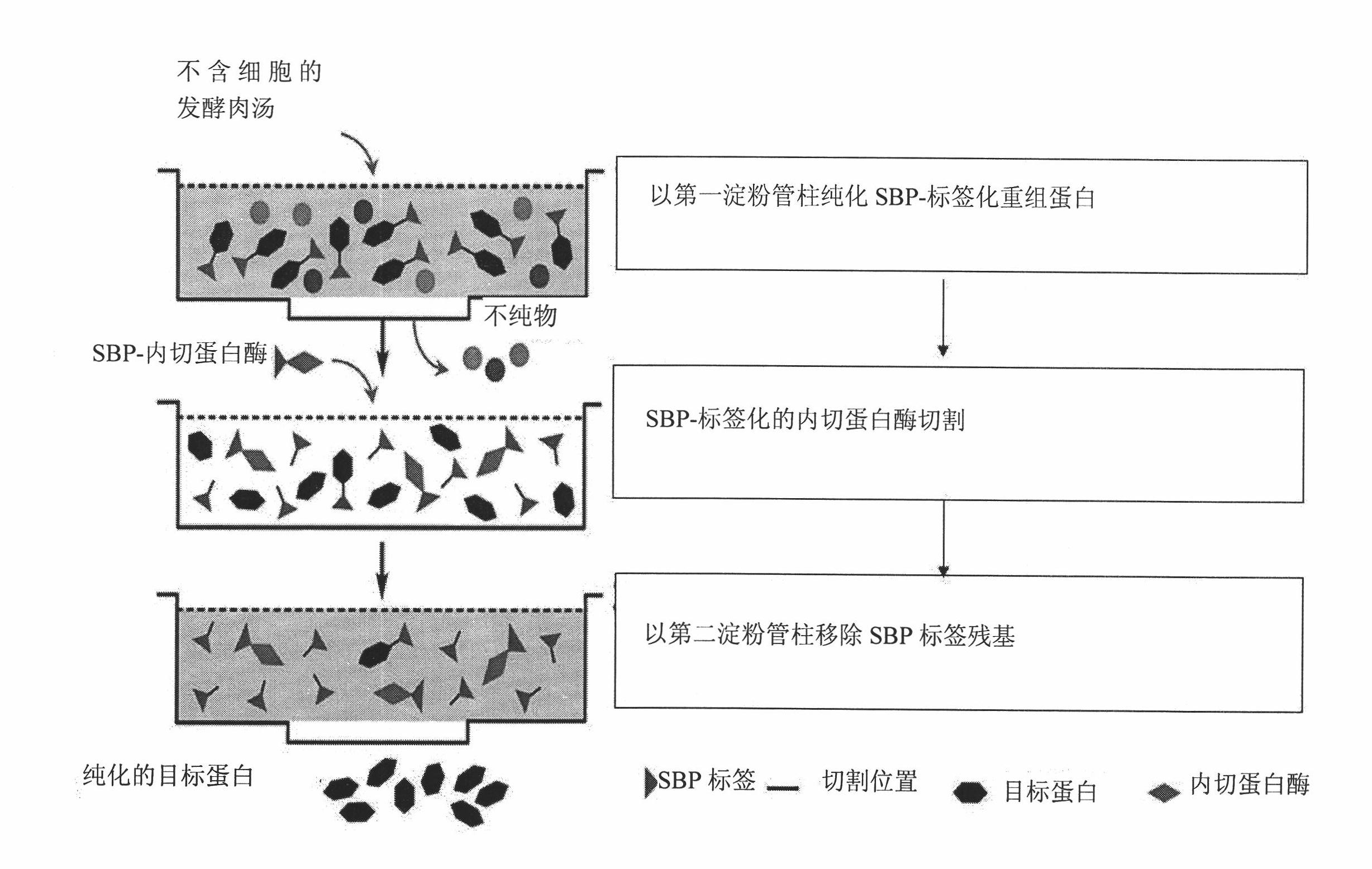
[0084] Example 1 Protein Purification
[0085] SBP-tagged recombinant protein construction and expression
[0086] The SBP gene is amplified by PCR and fused to the N-terminal of the target protein, which contains an endoprotease recognition site between the SBP and target protein genes. The fusion protein was replicated in the Pichia pastoris expression vector pPICZαA under the control of the AOX1 promoter and transformed into Pichia pastoris GS115 for expression. Pichia pastoris transformants carrying the SBP-target protein gene were cultured in BMGY medium for 24 hours. The cells were obtained by centrifugation and resuspension in BMMY containing 0.5% methanol. 0.5% methanol (0.5% v / v) was added every 24 hours to induce the expression of SBP-tagged recombinant protein. After 5 days of induction, the cells were removed by centrifugation and the cell-free broth was collected for subsequent purification. For subsequent purification methods, please refer to figure 1 and th...
Embodiment 2
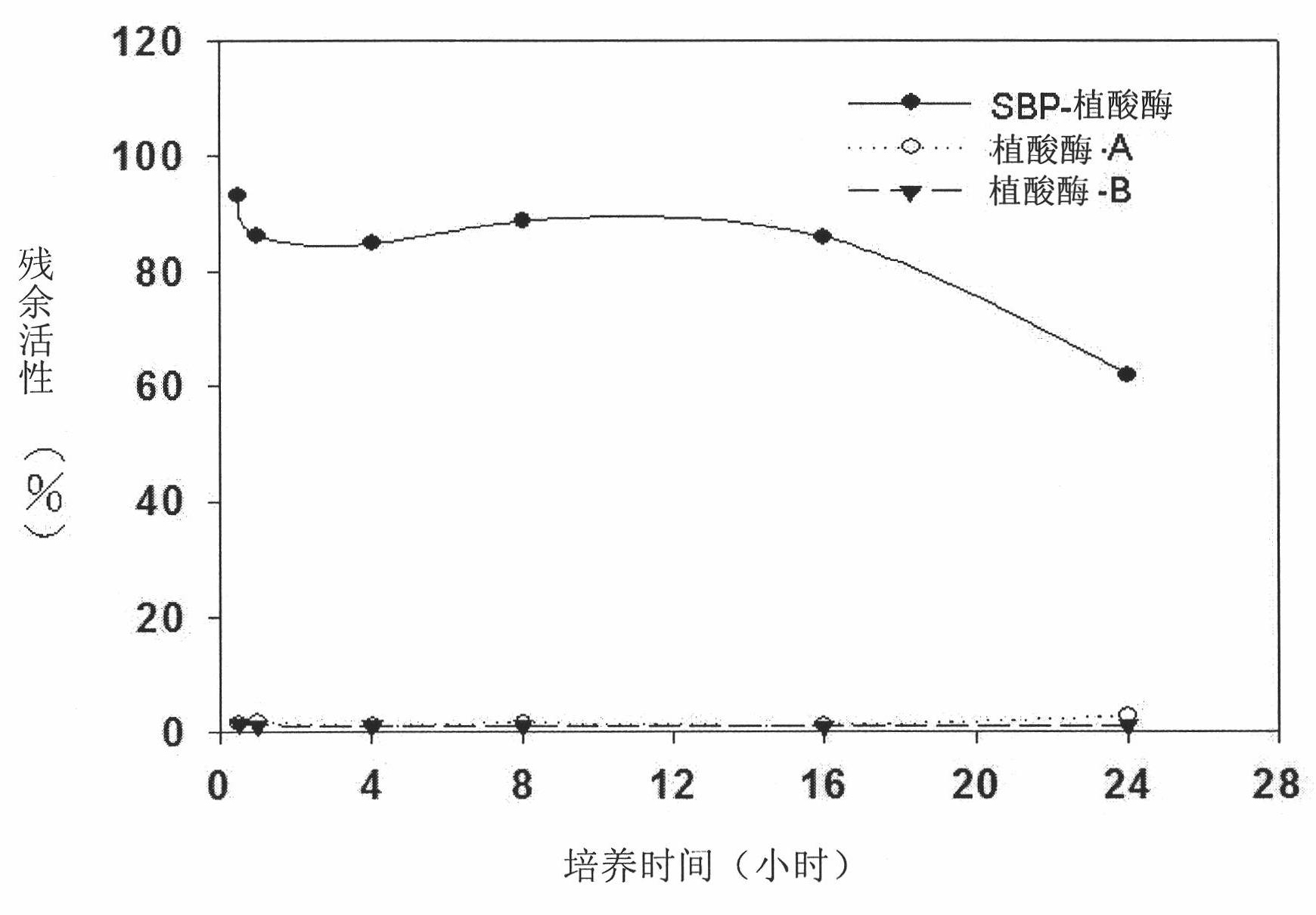
[0093] The comparison of embodiment 2 thermal stability
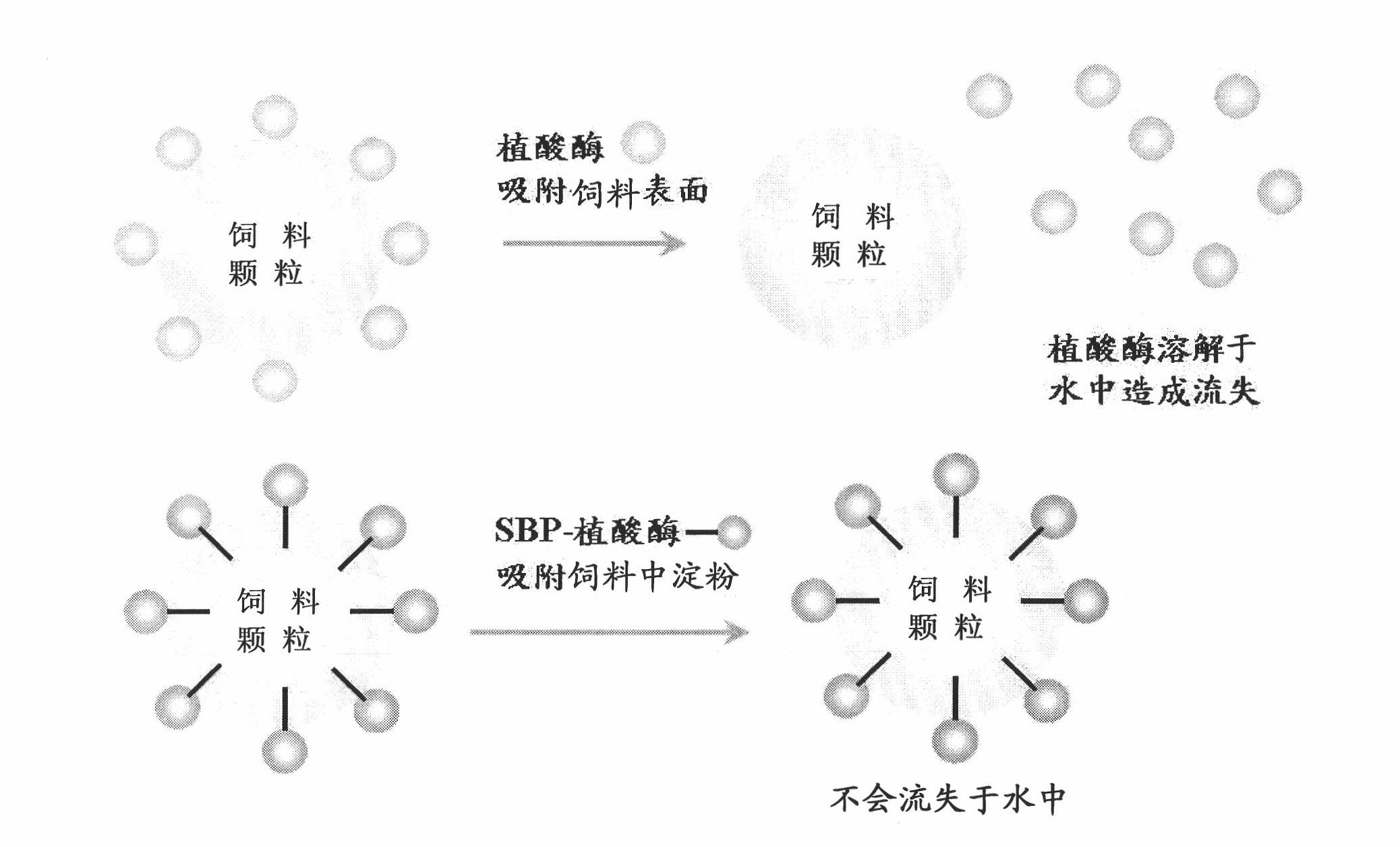
[0094] The SBP gene was amplified by PCR and fused to the N-terminus of the target enzyme. The lipase gene from R. oryzae, the xylanase gene from unpurified rumen fungal culture medium and the phytase gene from E. coli were all duplicated and fused to SBP. The fusion proteins such as SBP-lipase, SBP-xylanase and SBP-phytase are expressed by the above method and used in the present invention.
[0095] Thermostability of lipases from different sources
[0096] The lipase activity unit (U) is defined as when the sample releases 1 μmole of p-Nitrophenol per minute in 0.3 mM 4-Nitrophenyl palmitate (4-Nitrophenylpalmitate) at 30 ° C, pH Under the condition of value 7.0. SBP-lipase (52500 U / g) and F-AP15 lipase (32430 U / g, R. oryzae lipase purchased from Amano Enzyme Inc.) were mixed with 15% or 20% water content of fermented soybean flour to reach 100 U per gram. 100U / g lipase mix from different sources was weighed and p...
Embodiment 3
[0121] Example 3 Binding test of starch binding protein-enhanced green fluorescent protein (hereinafter referred to as SBP-eGFP) bound to different types of SBP matrices.
[0122] Binding assay of SBP-eGFP bound to different types of resins
[0123] 2 mg of unwashed branched starch, amylose resin (purchased from Bio-Rad Laboratories Inc., Hercules, California, U.S.), dextrin resin (purchased from GE Healthcare., Waukesha, U.S.) and sephadex (a kind of glucan Sugar commercial product, purchased from Sigma., Saint Louis, Missouri, U.S) was stirred together with SBP-eGFP in binding buffer (50 mM NaOAc, pH 5.5), its concentration was 0.3 mg / mL, and the overall volume was 200 μL. After stirring and incubation at 25°C for 3 hours, the samples were centrifuged. The suspension (unbound protein) and resin pellets (bound protein) were boiled and used for SDS-PAGE. The results of this binding test are shown in Figure 4A to Figure 4C .
[0124] Binding assay of SBP-eGFP bound to algi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com