Method for constructing RNA (Ribonucleic Acid) interference vector by directly annealing multi-primers
A technology of RNA interference and multiple primers, applied in the field of biological gene cloning and expression technology, can solve the problems of not being suitable for high-throughput, time-consuming, and heavy workload, so as to avoid base synthesis errors, save time and cost, and reduce the probability of errors Reduced effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
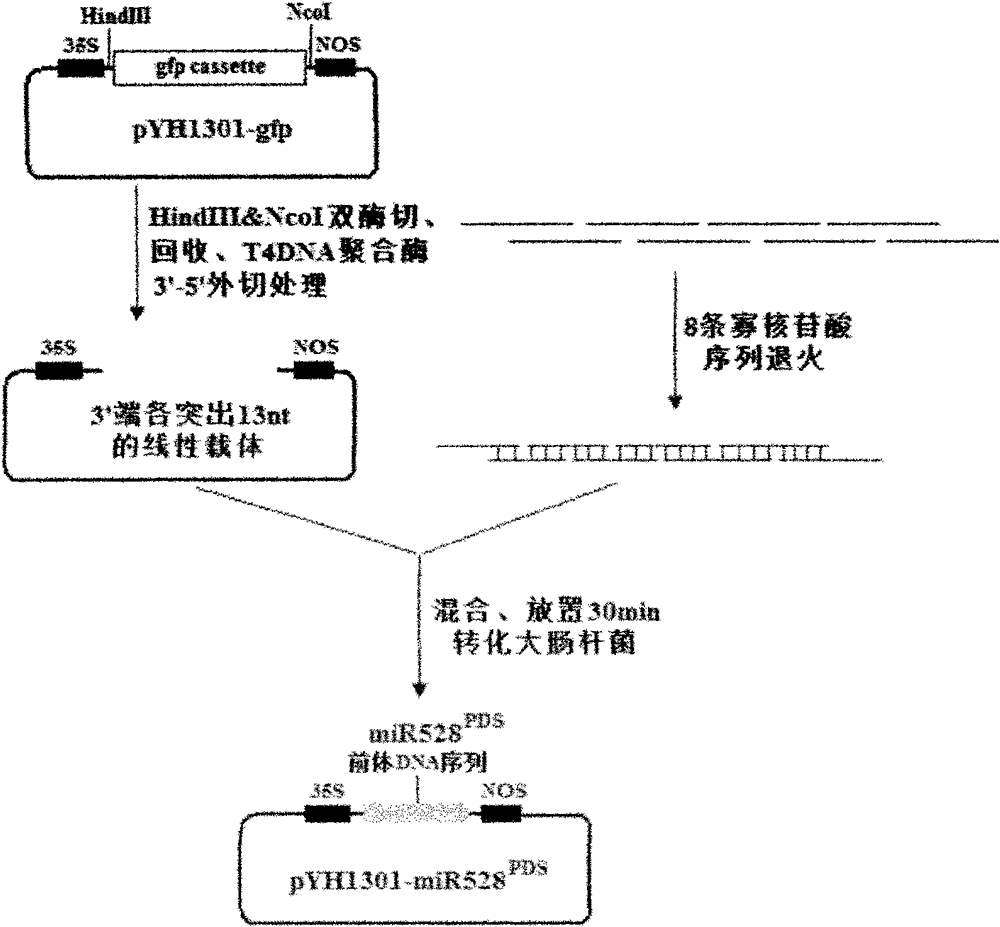
[0031] Utilize the present invention, construct the plant artificial microRNA (amiRNA) expression vector (specific process sees figure 1 ).
[0032] 1) Transformation of the carrier
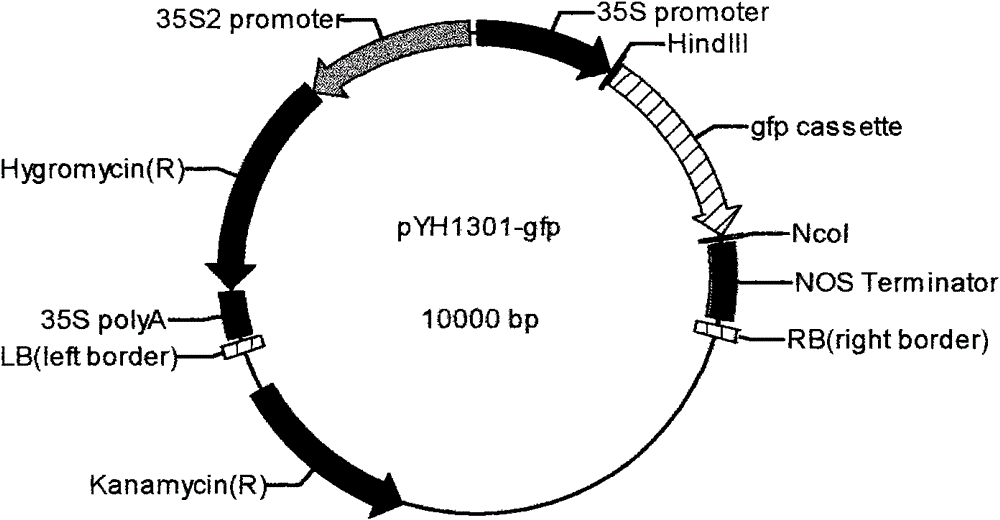
[0033] In this experiment, the plant binary vector pCAMBIA1301 was selected as the starting vector, and the vector was digested with EcoRI and BstEII and then ligated with the following fragments: 35S promoter-HindIII restriction site-gfp expression unit-NcoI restriction site (details The sequence is attached), to obtain the universal plant RNAi expression vector pYH1301-gfp (see figure 2 ).
[0034] 2) Design and annealing of oligonucleotide sequence
[0035] According to the rice miR528 precursor sequence reported in the literature (PLoS One.2008Mar 19; 3(3):e1829.), design the sequence that can transcribe the small RNA that interferes with the pds gene, and synthesize a set (8 in this case) of length It is an oligonucleotide sequence of about 50nt (the detailed sequence is attached).
[...
Embodiment 2
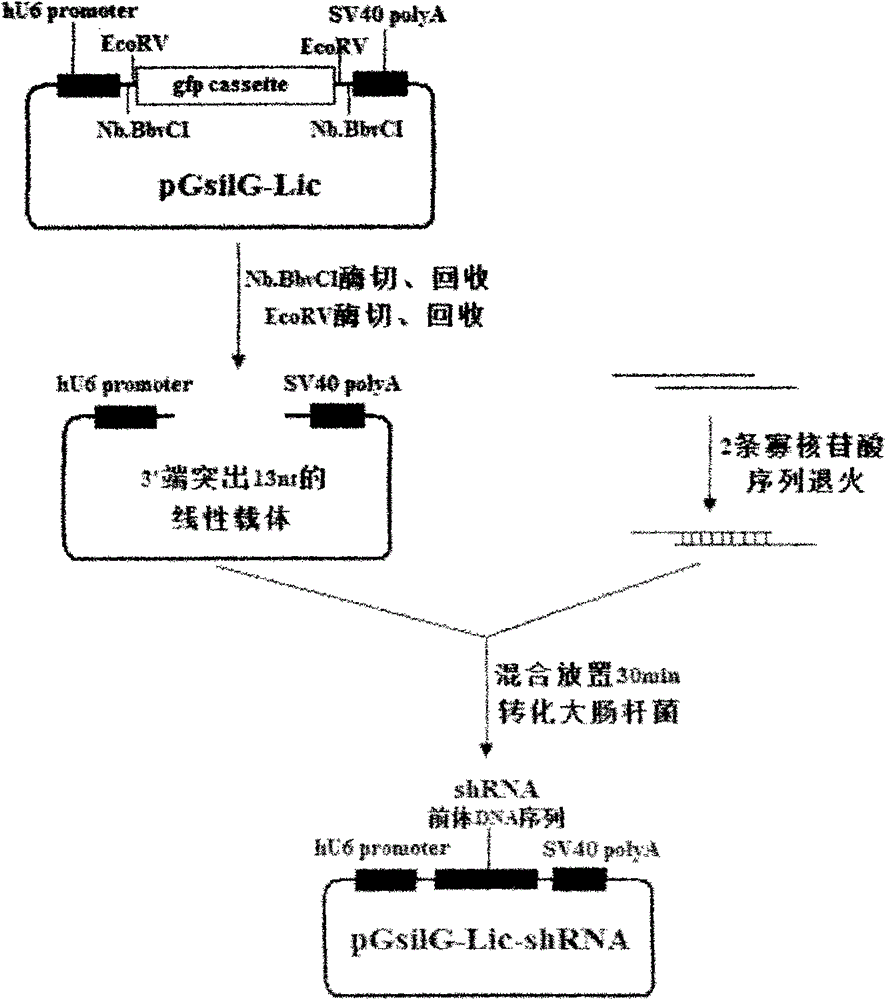
[0040] Utilize the present invention to construct the animal shRNA expression vector with mammalian interference vector pGenesil-1 (Wuhan Jingsai Bioengineering Co., Ltd.) as the backbone and GCTGTACTCCCCTTACATTA as the interference target sequence (see image 3 ).
[0041] 1) Transformation of the cloning vector
[0042] To analyze the vector sequence, first select the mammalian interference vector pGenesil-1 as the starting vector, linearize it with BamH I and Xho I double enzyme digestion, and fragment "Nb.BbvCI restriction site--front annealing sequence--EcoRV Restriction site--gfp expression unit--EcoRV restriction site--rear-end annealing sequence--Nb.BbvCI restriction site" connection (detailed sequence is attached) to obtain a general-purpose gene that can be used to construct animal shRNA expression vectors Type vector pGsilG-Lic ( Figure 4 );
[0043] 2) Design and annealing of oligonucleotide sequence
[0044] According to the principle in step 2) of the specif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com