Application of chromatin remodeled protein 4A (CHMP4A) in enhancing stability and transcriptional activity of hypoxia-inducible factor 1alpha (HIF-1alpha)
A hypoxia-inducible factor and chromatin technology, applied in the fields of application, recombinant DNA technology, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
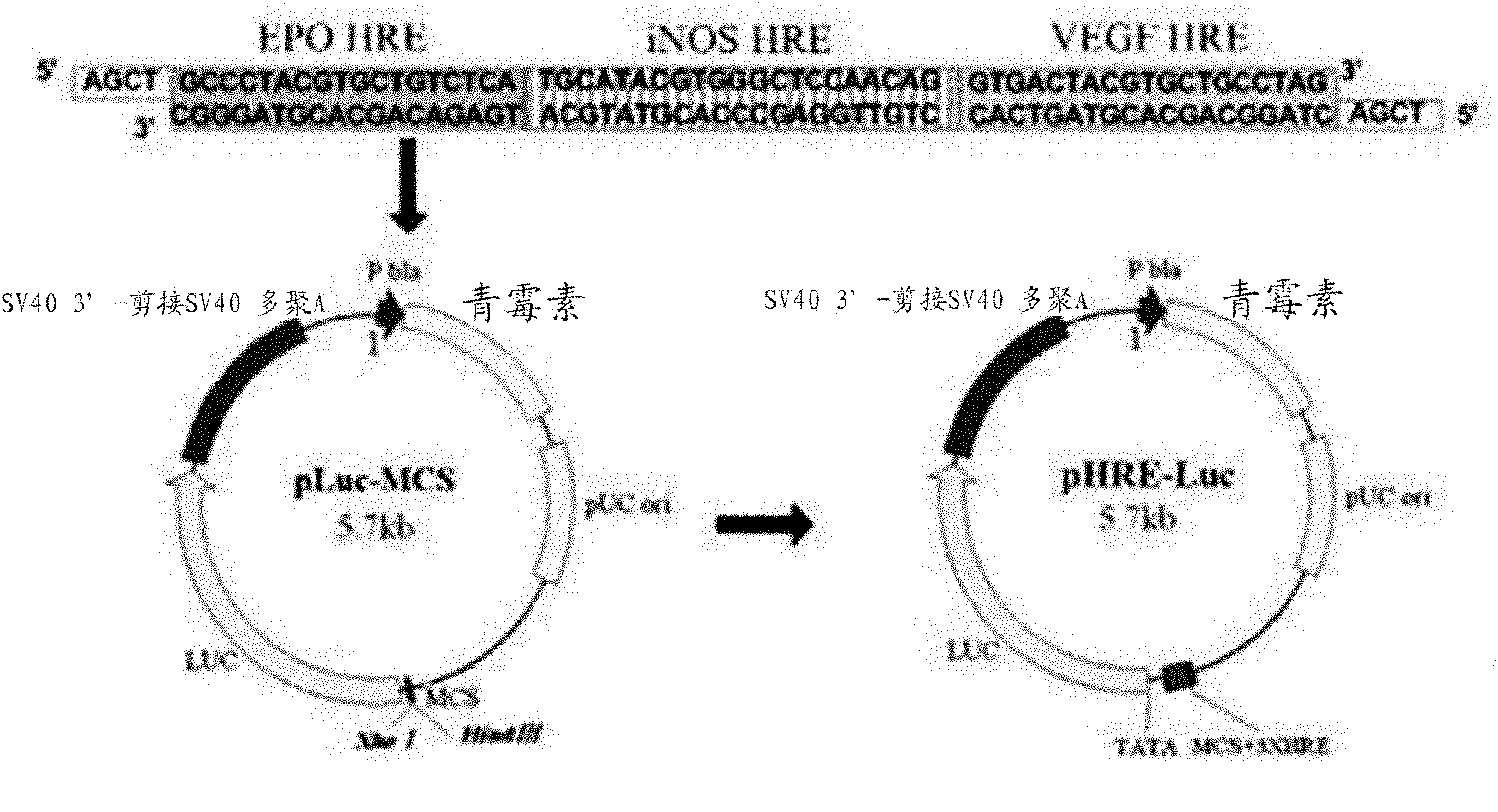
[0039] The construction of embodiment 1 reporter gene plasmid (pHRE-LUC)
[0040] The HRE sequence of the promoter region of three genes consisting of erythropoietin (Erythropoietin, EPO), inducible nitric oxide synthase (iNOS), and vascular endothelial growth factor (VEGF) 3×HRE sequences were synthesized by Shanghai Sangon. Sense strand (5'-AGCTGCCCTA CGTGCTGTCT CATGCATACG TGGGCTCCAA CAGGTGACTA CGTGCTGCCT AG-3', containing sticky-end sequence of HindIII) and antisense strand (5'-TCGACTAGGC AGCACGTAGT CACCTGTTGG AGCCCACGTA TGCATGAGAC AGCACGTAGG GC-3', containing sticky-end sequence of Xho I ) annealed into a double strand and connected to the pLUC-MCS empty vector cut by HindIII and Xho I, thereby obtaining the reporter gene plasmid pHRE-LUC (such as figure 1 shown).
Embodiment 2HI
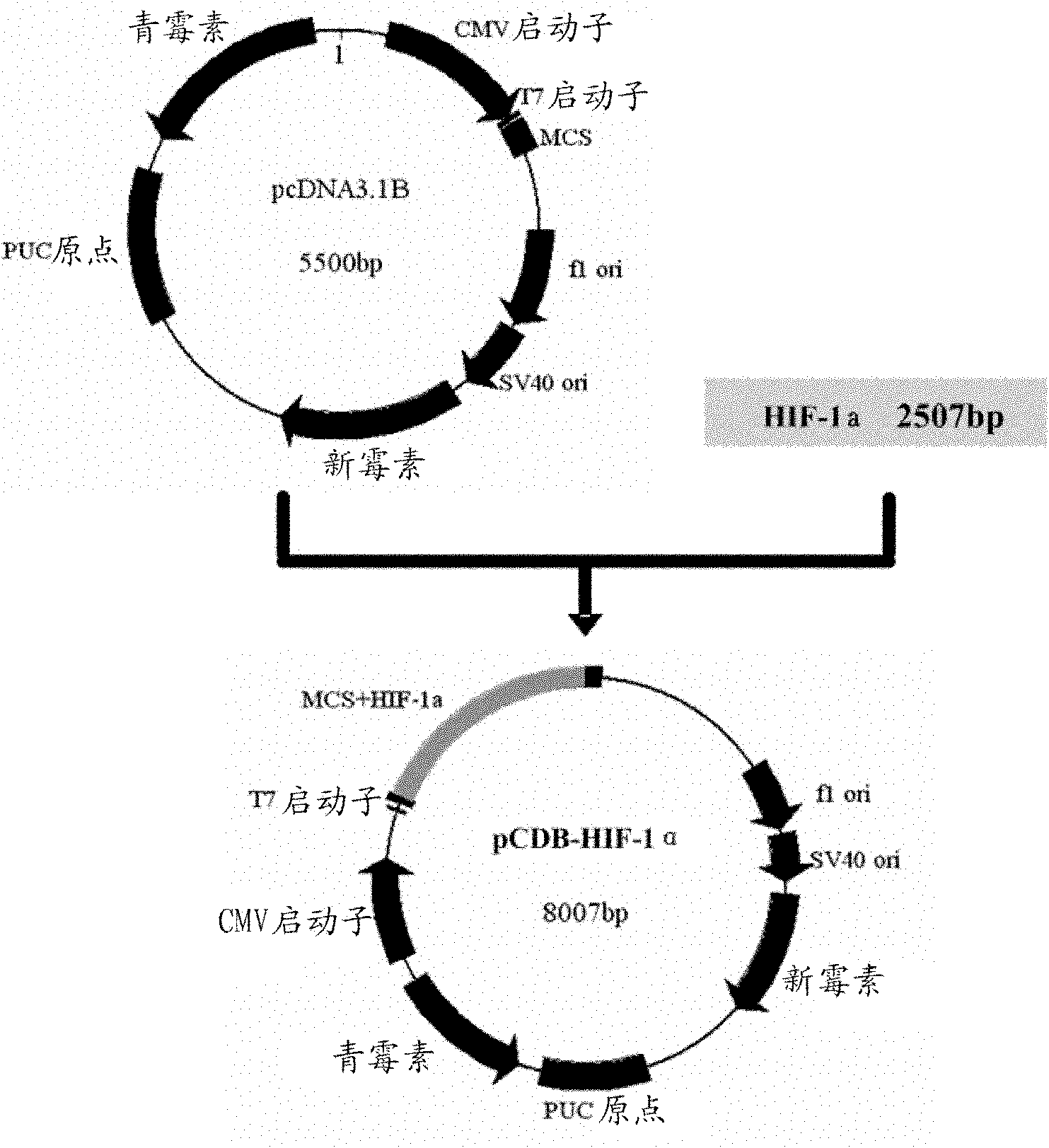
[0041] Construction of embodiment 2 HIF-1α eukaryotic expression plasmid
[0042] According to the full-length HIF-1α sequence (GenBank NM_001530), the upstream primer (5-GAAGACATCG CGGGGACC-3) and the downstream primer (5-GGGGTACCGAAAAAAGCTCAGTTAACTTGATC-3) were PCR-amplified from the human normal fetal liver cDNA library to a full-length 2507 bases , connected to the T-easy carrier after gel recovery. Then use Not I single enzyme digestion and connect to pcDNA3.1 / myc-His(-)B (pCDB) with isoenzyme cutting site, thereby obtain HIF-1α eukaryotic expression plasmid pCDB-HIF-1α( figure 2 ). The size of the plasmid conforms to the calculated value ( image 3 ).
Embodiment 3
[0043] The detection of embodiment 3 dual-luciferase reporter system
[0044] Hela cells were routinely subcultured in DMEM medium (HyClone, SH30022.01B) containing 10% fetal bovine serum (HyClone, SH30084.03) at 37°C, 5% CO 2 cultured in an incubator. According to 1.1×10 6 The density of cells / ml is inoculated in 96-well plates, 100 μl per well, and cultured in an incubator for 18 hours to 24 hours, so that the cell density reaches 40% to 60%, ready for transfection. According to the instructions, 50ng of pHRE-LUC, 5ng of pRL-TK-LUC and 50ng of the gene to be screened or the control plasmid were co-transfected into Hela cells using VigoFect cationic transfection reagent (Vigolas Biotechnology (Beijing) Co., Ltd.). Cells transfected with pcDNA3.1 / myc-His(-)B empty vector were used as blank control. Cells transfected with pCDB-HIF-1α and ING4 gene (ING4 connected in PCDB vector, as a positive control, similar to HIF-1a) were used as positive controls for activating the expre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com