Non-competitive internal controls for use in nucleic acid tests
A non-competitive, internal control technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as inability to use multiple analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] Preparation of the MET IC DNA insert sequence
[0089] Genomic DNA was extracted from MET samples. DNA inserts were prepared by PCR on genomic DNA using the fragment primers in Table 1. The following sequence is the sequence (SEQ ID NO: 37) of the MET IC PCR product (213 bp). XhoI and SpeI restriction enzyme sites are underlined in bold.
[0090] 1 AGTAGTC CATGTGCA GGGATCCTGA CACGGTACTG
[0091] TCATCAG GTACACGT CCCTAGGACT GTGCCATGAC
[0092] 41 GAGGCAGGCA GGGCCGCCAT AAGAGCCATA GAGGAGGTTG
[0093] CTCCGTCCGT CCCGGCGGTA TTCTCGGTAT CTCCTCCAAC
[0094] 81 AGGGTGTTGT GACGCCCTTT GATATCTGCT CCGCAGCATC
[0095] TCCCACAACA CTGCGGGAAA CTATAGACGA GGCGTCGTAG
[0096] 121 AAAGCCAGAG ACAAATTACC CCTGGATAGG CCCCACCACG
[0097] TTTCGGTCTC TGTTTAATGG GGACCTATCC GGGGTGGTGC
[0098] 161 AACCACCCCT ACTGCCCGAG CCTGAAGGAG GTGCTCGGTG
[0099] TTGGTGGGGA TGACGGGCTC GGACTTCCTC CACGAGCCAC
[0100] 201AA CGA CGA
[0101] TT GCT GCT
Embodiment 2
[0103] Purified MET IC DNA Insert Sequence
[0104]The following sequence is the purified 195 bp dsDNA sequence (SEQ ID NO: 38) following restriction enzyme digestion at the sites identified above:
[0105] 1 TCGAGCATGT GCAGGGATCC TGACACGGTA CTGGAGGCAG
[0106] 41 GCAGGGCCGC CATAAGAGCC ATAGAGGAGG TTGAGGGTGT
[0107] 81 TGTGACGCCC TTTGATATCT GCTCCGCAGC ATCAAAGCCA
[0108] 121 GAGACAAATT ACCCCTGGAT AGGCCCCACC ACGAACCACC
[0109] 161 CCTACTGCCC GAGCCTGAAG GAGGTGCTCG GTGAA
Embodiment 3
[0111] MET IC DNA transcript sequence
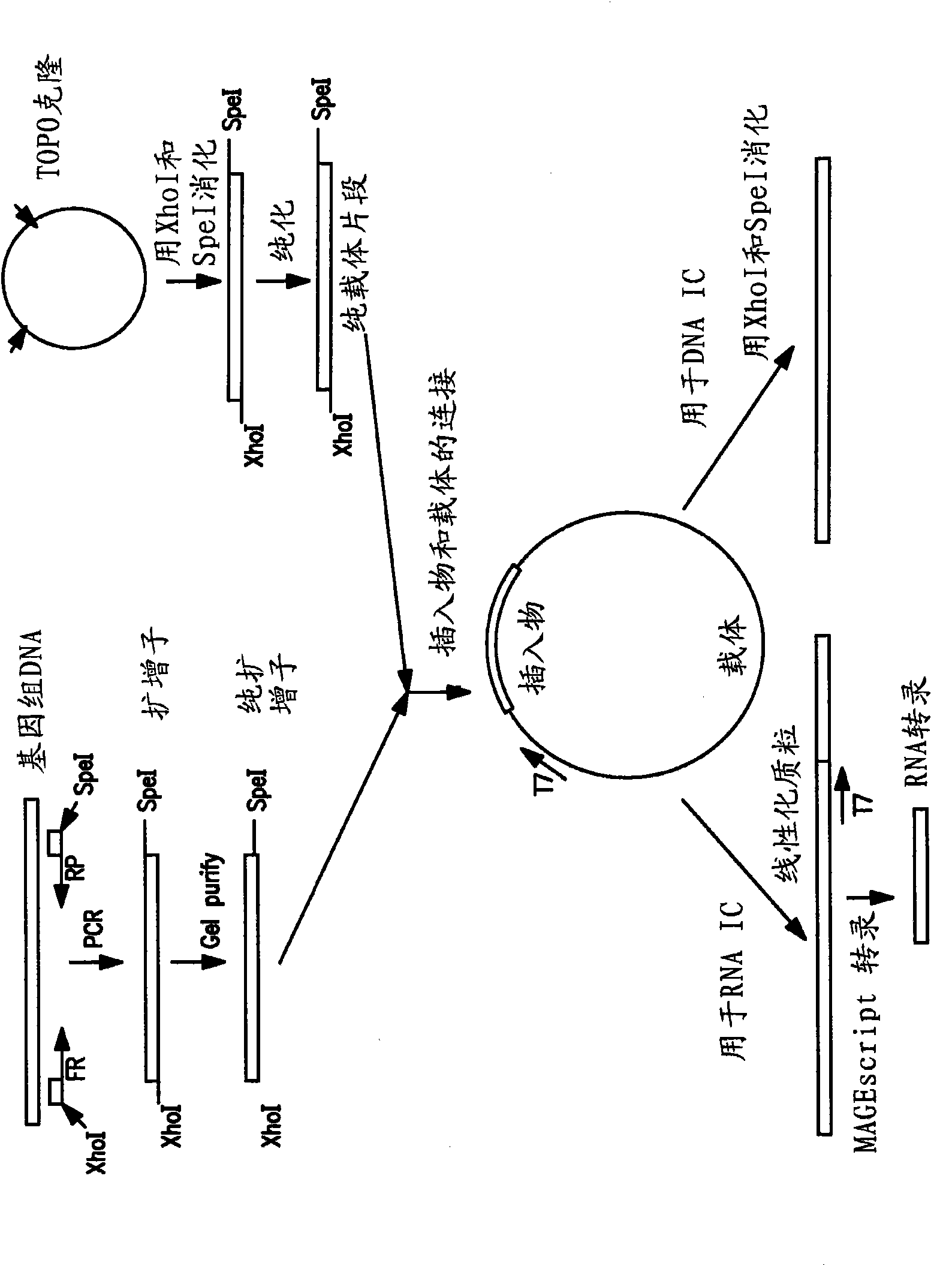
[0112] A plasmid was prepared by ligating the purified MET IC DNA insert sequence of Example 2 into a purified vector fragment and adding the T7 promoter sequence. Purified vector fragments were isolated from Cloning vector (Invitrogen, Carlsbad, California). Plasmids are formed by matching the XhoI and SpeI cohesive ends of the DNA insert and the vector. attached figure 1 A schematic diagram of the cloning process is shown.
[0113] The resulting plasmid was linearized with XhoI and SpeI to generate the following 247bp MET ICDNA transcript sequence. The vector sequence is underlined in bold (SEQ ID NO: 39):
[0114] 1 CTC GAGCATGTGC
[0115] 41 AGGGATCCTG ACACGGTACT GGAGGCAGGC AGGGCCGCCA
[0116] 81 TAAGAGCCATAGAGGAGGTTGAGGGTGTTGTGACGCCCTT
[0117] 121 TGATATCTGC TCCGCAGCAT CAAAGCCAGA GACAAATTAC
[0118] 161 CCCTGGATAG GCCCCACCAC GAACCACCCC TACTGCCCGA
[0119] 201 GCCTGAAGGA GGTGCTCGGT GAACTAGT
[0120] 241
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com