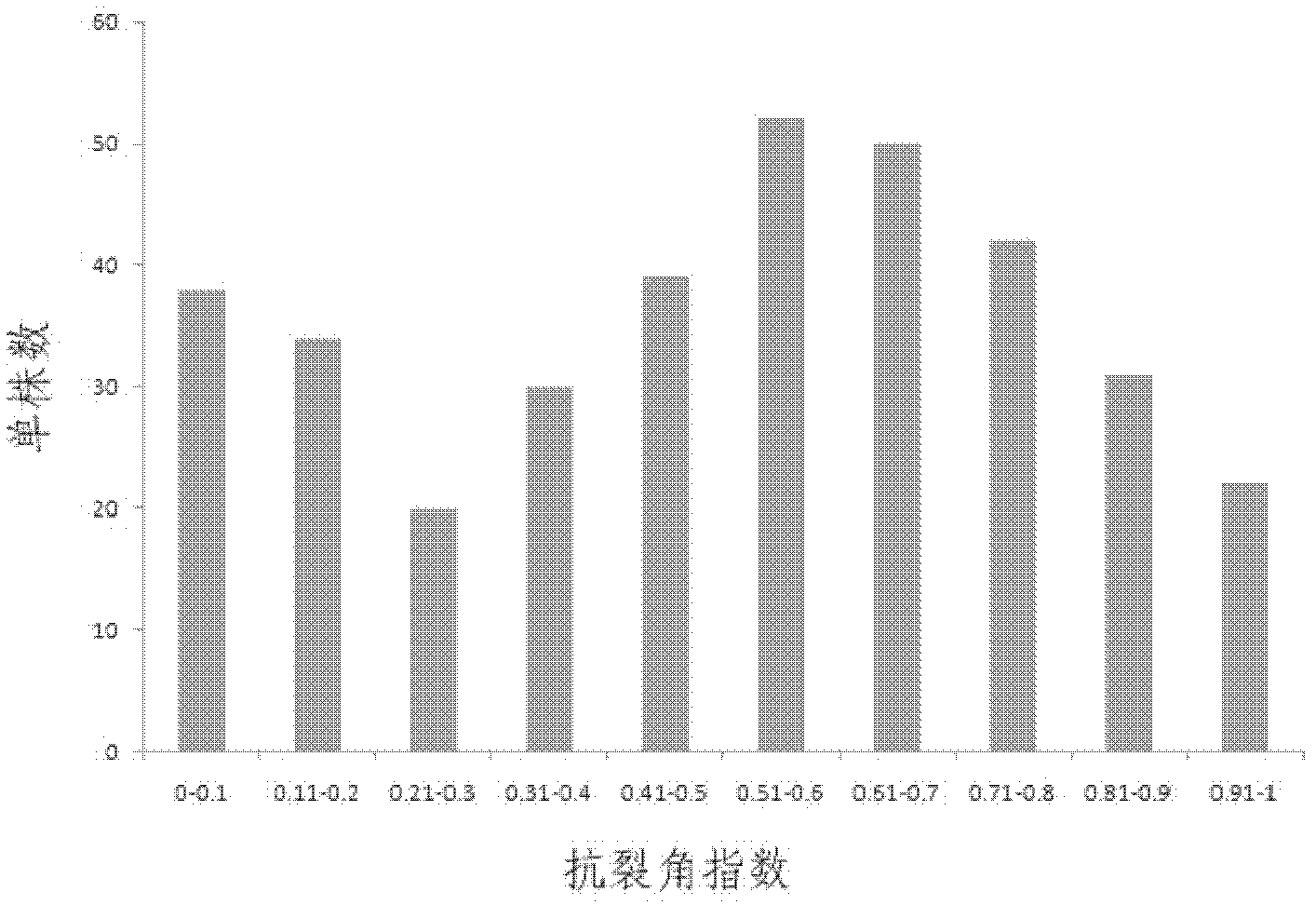Structure and major gene locus Psr9 of pod shattering resistance character of rape and application thereof
A major gene and gene locus technology, applied in the fields of molecular biology and genetics and breeding, can solve the problems of quantitative traits being easily affected by environmental conditions, long cycle, poor selection effect, etc., achieving convenient and rapid detection and high cost. , the effect of improving the selection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] A method for preparing the main gene locus Psr9 of the anti-cracking angle trait of rapeseed, the steps of which are:
[0041] a) The population used in this example is the F2 offspring of high and low cleavage-resistant parents (zy72360 with a cleavage-resistant index of 0.94 and R1 with a cleavage-resistant index of 0.06). The anti-cracking angle index of parents, F1 and F2 seeds was determined by random collision method. The results of the data distribution of the anti-crack angle index of the F2 generation segregation population showed that the performance distribution of the anti-crack angle trait was a continuous distribution, but the variation distribution did not show a normal distribution, which proved that the crack angle resistance trait was a quantitative trait and there was a main effect gene locus (see figure 1 ). The combination of zy72360 and R1 was used to cross rapeseed with high and low anti-cracking angles, and the F1 generation was self-crossed to...
Embodiment 2
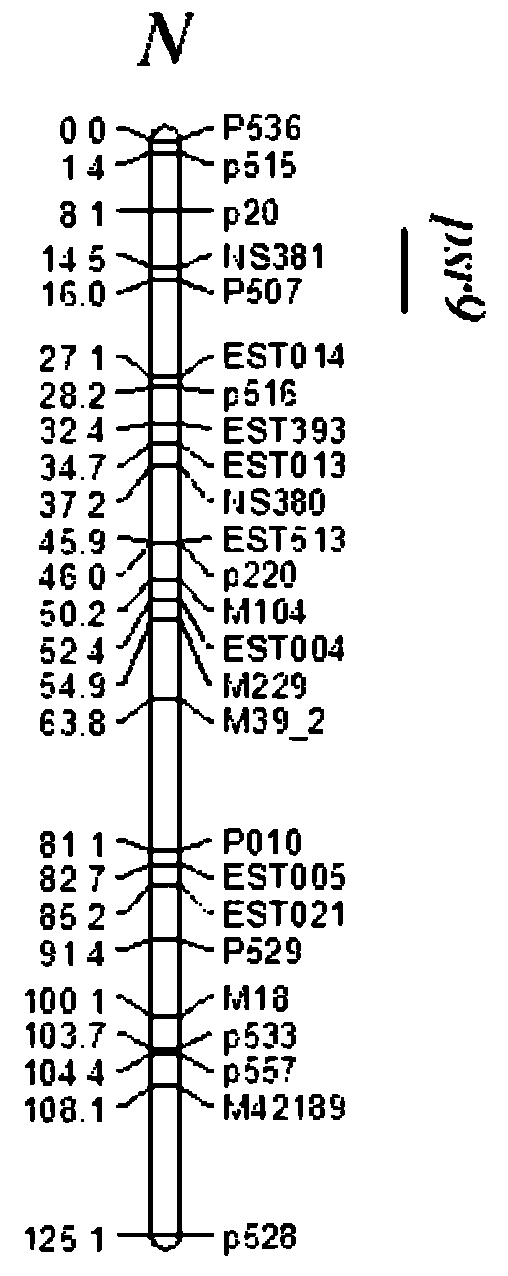
[0061] The application of the main effect gene locus Psr9 of rapeseed's high cracking horn resistance trait breeding, and the P507 sequence of the A9 linkage group where it is located in the rapeseed high cracking horn trait breeding, the steps are:
[0062] a) The population used in this example is the F2 offspring of high and low cleavage-resistant parents (zy72360 with a cleavage-resistant index of 0.94 and R1 with a cleavage-resistant index of 0.06). The F2 generation materials are planted in the field, and numbered according to individual plants. In this embodiment, 400 individual plants are numbered, and the numbers are 001-400.
[0063] b) Extract DNA from 400 individual plants of the F2 generation numbered 001-400 at the seedling stage by taking a small piece of leaf. The extraction method refers to the DNA extraction method in Example 1.
[0064] c) Using the marker P507 primer of Psr9, the main gene locus of the anti-cracking angle trait of rapeseed, to perform PCR a...
Embodiment 3
[0069] A structure highly correlated with anti-splitting angle traits in rapeseed, application of fruit petal and placenta frame combined area index in breeding for anti-splitting angles. The steps are:
[0070] a) Apply in conjunction with Example 2. For the 89 F2 generation plants screened by molecular markers in Example 2, take siliques at the late stage of silique development, observe and measure the combined area index of the fruit petals and the placenta frame through a dissecting microscope .
[0071] b) The results show that among the 89 F2 generation plants screened by molecular markers, 56 (63%) plants have a combination area index of fruit petals and placenta frame ≥ 2mm 2 . The anti-cracking angle test results after the seeds were harvested showed that among the 56 plants screened by molecular markers and the combined area index of fruit petals and placenta frame, 48 (accounting for 86%) had an anti-cracking angle index above 0.80. That is to say, 86% of the obt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com