Aptamer capable of identifying hepatitis C virus (HCV) nonstructural 5A (NS5A) protein, derivatives thereof and screening method and use thereof
A nucleic acid aptamer and screening method technology, applied in the field of genetic engineering, can solve the problems of patient interruption of treatment, expensive treatment, and serious side effects of combination therapy, and achieve the effects of inhibiting HCV replication, specifically inhibiting HCV infection, and being easy to preserve.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
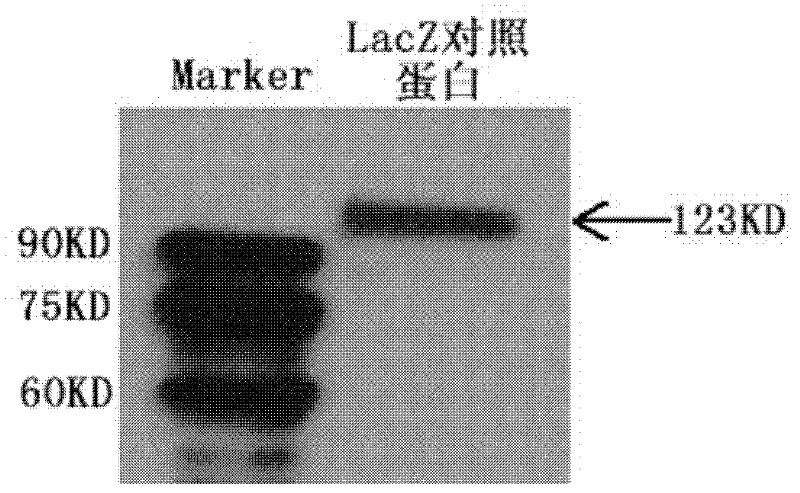
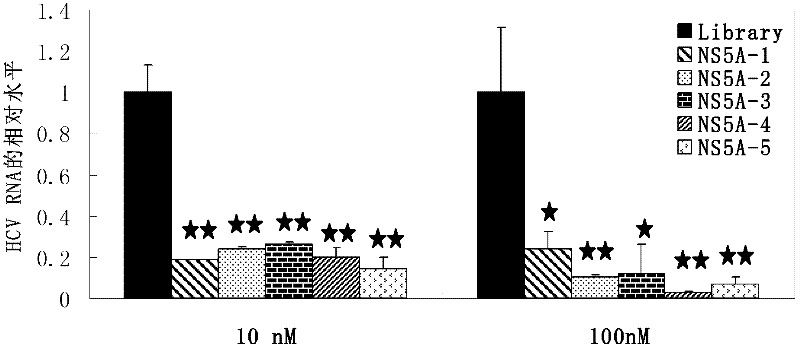
[0053] Example: nucleic acid aptamers capable of recognizing HCV NS5A protein, derivatives of nucleic acid aptamers, screening methods and applications thereof.
[0054] 1. Preparation of related proteins.
[0055] 1.1 Preparation of HCV NS5A protein (ie target protein) with histidine tag.
[0056] 1.1.1 Amplification of the gene encoding HCV NS5A protein
[0057] Using the pJFH1 plasmid as a template for PCR amplification, the NS5A gene was amplified by PCR with the first primers consisting of the following primers 1 and 2 (PCR reagents were purchased from Roche, and 100 microliters of PCR tubes were purchased from Eppendorf):
[0058] Primer 1 (upstream primer):
[0059] 5'-CGCGCCATATGTCCGGATCCTGGCTCCGCGAC-3';
[0060] Primer 2 (downstream primer):
[0061] 5'-CTGCAGAAGCTTTCAGCAGCACACGGTGGTATCG-3';
[0062] The 5' ends of the upstream and downstream primers were respectively introduced with Nde I restriction site and HindIII restriction site;
[0063] The PCR amplifica...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com