Real time in situ characterization method for single biomolecular reaction
A biomolecular and reaction technology, applied in biological testing, individual particle analysis, particle and sedimentation analysis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
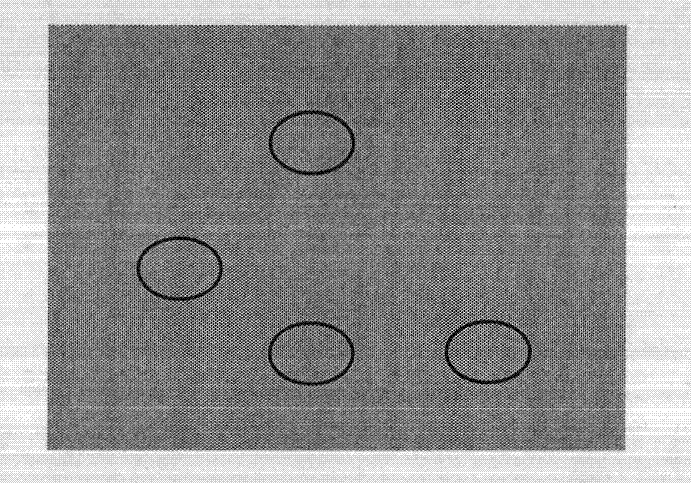
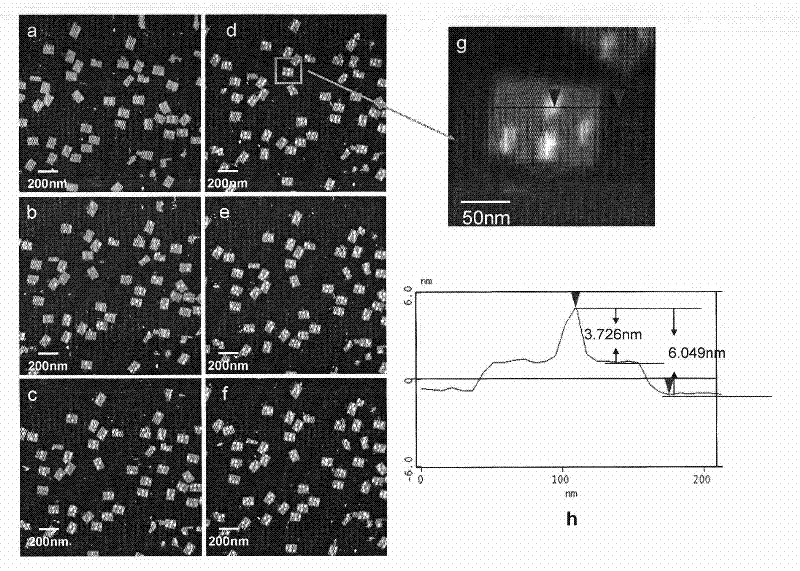
[0035] 1. The DNA origami designed in this example utilizes single-strand self-assembly of M13mp18 DNA and staples to form a rectangular pattern with a length and width of 100nm and 70nm, respectively, such asfigure 1 As shown, at four positions on the DNA origami, the ends of the staple strands are biotinylated for binding to streptavidin.
[0036] Preparation of specific biotinylated square DNA origami:
[0037] M13mp18 DNA single strands and staple single strands (including biotinylated staple single strands) were mixed at a molar concentration of 1:10 and placed in 1×TAE / Mg 2+ Buffer system (Tris 40mM, acetic acid 20mM, EDTA 2mM, MgCl 2 12.5mM, and the pH value is 8), the total volume is 61uL, and then placed on the PCR instrument and annealed from 95°C to 20°C at an annealing speed of 0.1°C / 10s, and stored at 4°C after the reaction is completed.
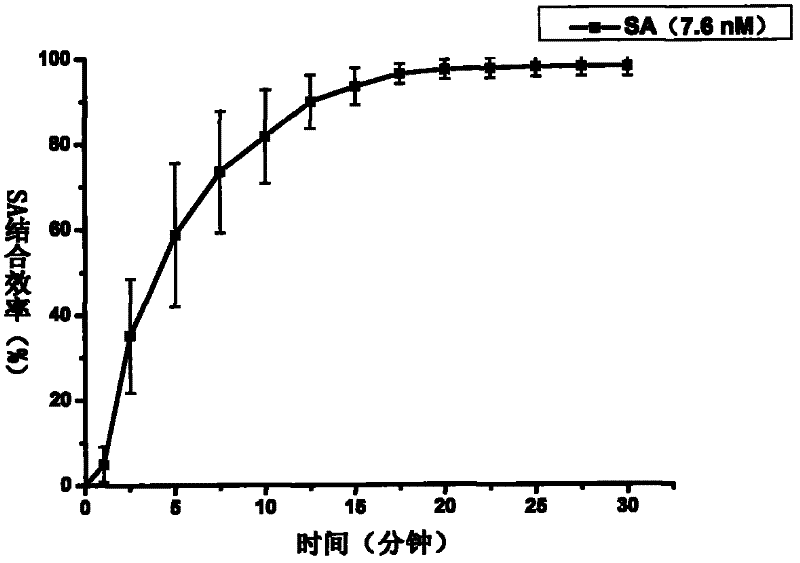
[0038] 2. Real-time in situ detection of biotin dynamic reaction process between streptavidin and biotin-modified DNA origam...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com