A dna sequencing method for shortening dna template and its application
A DNA sequencing and template technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of poor sequencing accuracy and achieve high accuracy, high sequencing accuracy, and improved read length.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
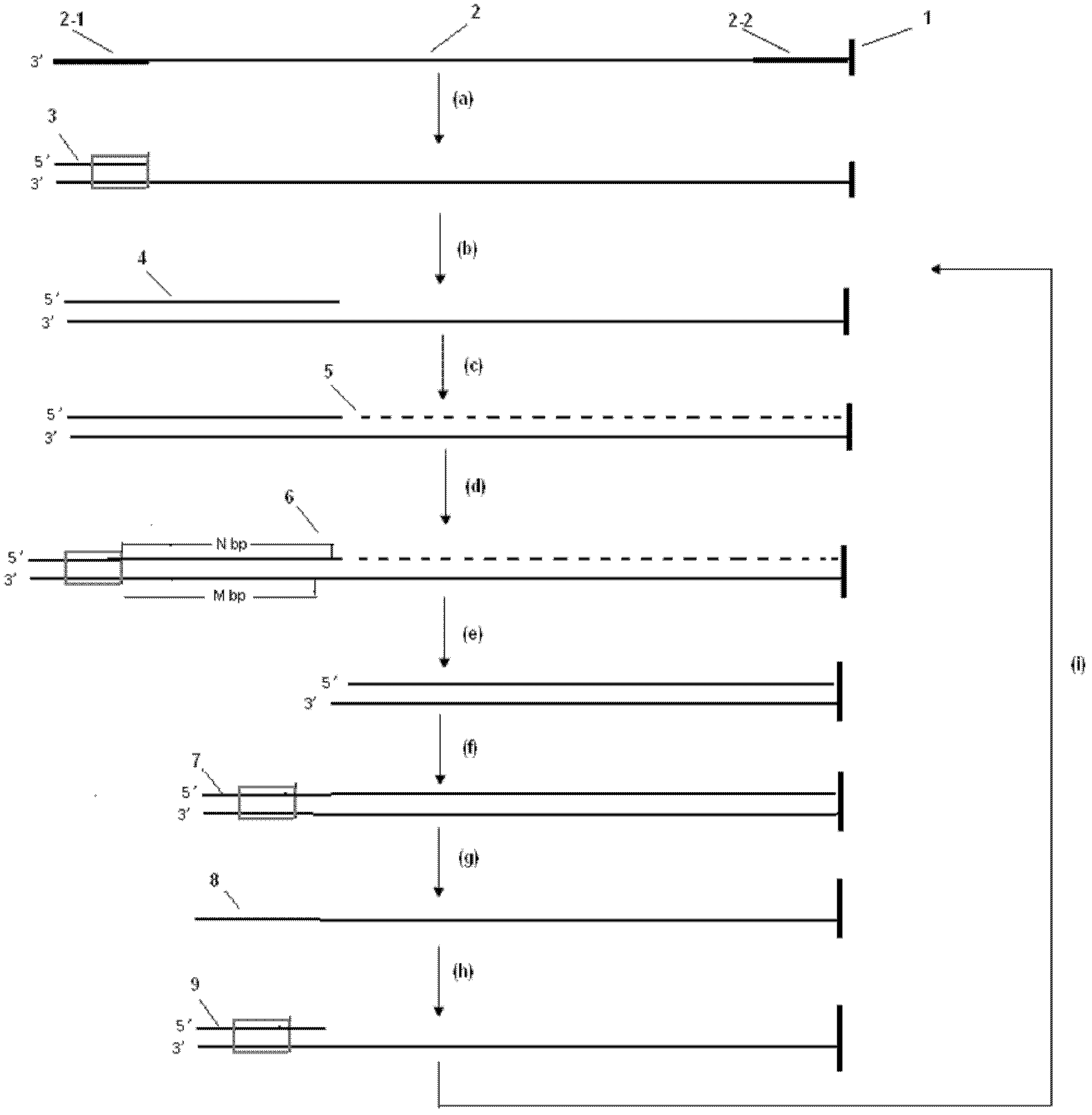
[0056] Example 1: Determination of human genome by ligation sequencing of shortened DNA template
[0057] (1) After the human genome was treated with Fok I enzyme at 37°C for 2 hours, it was ultrasonically broken into fragments with a size of 50-1000 bases, and these fragmented nucleic acid sequences were connected with a pair of universal linkers under the action of ligase Ligation (assumed to be 20 bases), wherein the oligonucleotide sequence of one universal linker is completely complementary to the sequence of the amplification primer, and the oligonucleotide sequence of the other linker contains the Mly I enzyme recognition sequence, and This specific region is located at the very beginning of template sequencing.
[0058] (2) The fragmented nucleic acid sequences connected by these tethers and the complementary sequences of the immobilized linkers are placed on microbeads to perform emulsion parallel PCR reaction to amplify the fragmented whole human genome. And these m...
Embodiment 2
[0071] Example 2: Determination of Escherichia coli genome by extension sequencing method of shortened DNA template
[0072] (1) After the E. coli genome was treated with Mme I enzyme at 37°C for 2 hours, it was ultrasonically broken into fragments with a size of 50-1000 bases, and these fragmented nucleic acid sequences were connected with a pair of universal ligation under the action of ligase The oligonucleotide sequence of one of the universal linkers is completely complementary to the sequence of the amplification primer, and the oligonucleotide sequence of the other linker contains the recognition sequence of the Mme I enzyme. And the specific region is located at the initial position of the template sequencing.
[0073] (2) The fragmented nucleic acid sequences connected by these tethers and the complementary sequences of the immobilized linkers are placed on microbeads to carry out emulsion parallel PCR reaction to amplify the fragmented Escherichia coli genome. And t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com