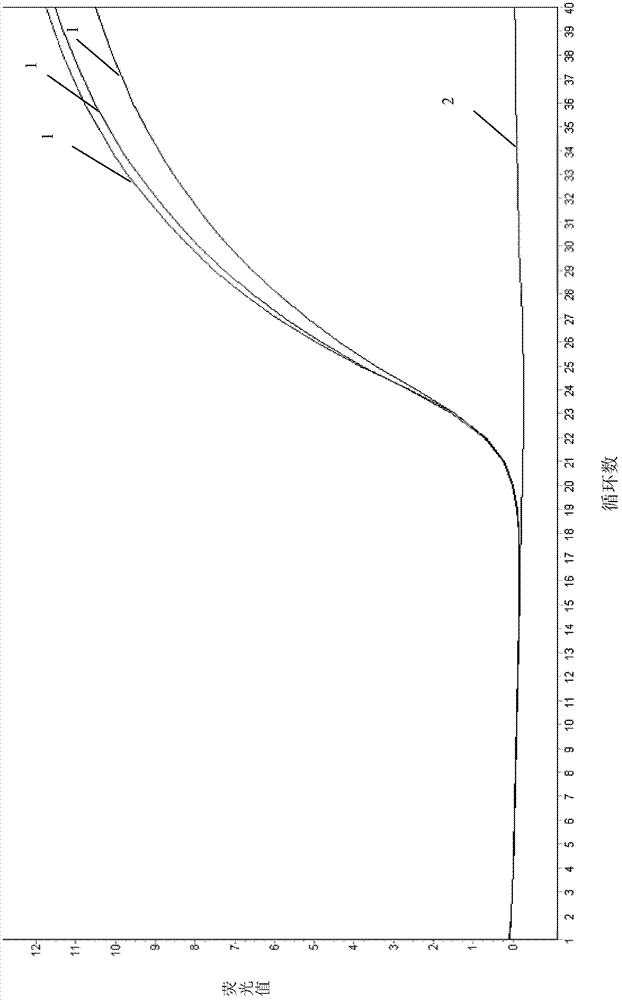
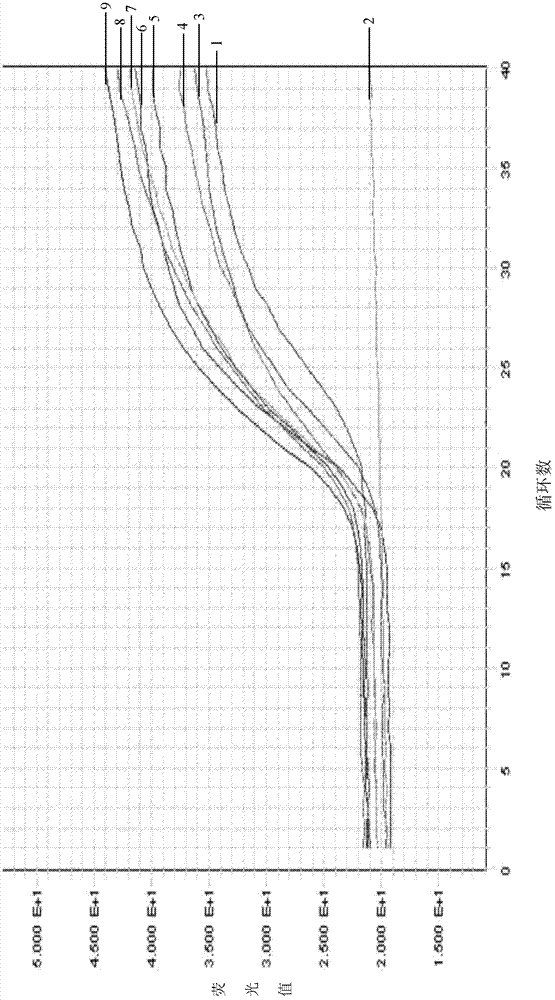
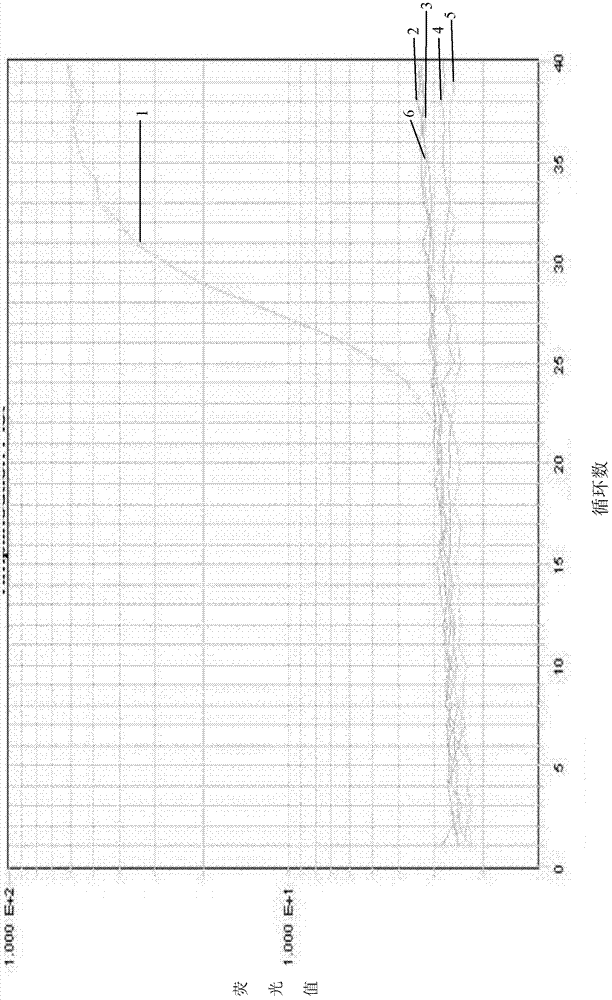
Real-time fluorescence PCR (polymerase chain reaction) kit and oligonucleotide sequence for detecting swine dysentery
A nucleotide sequence, oligonucleotide technology, applied in fluorescence/phosphorescence, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of lack of satisfactory detection methods, high detection conditions, and difficult laboratory requirements. Detection requirements and other issues to achieve the effect of improving specificity, reducing workload and improving work efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Design and synthesis of Taqman probes and primers
[0050] According to the relevant gene sequences of Treponema hyodysenteriae registered in Genbank, the DNAMAN software was used for comparison and analysis, and the highly conserved regions of the nox gene were selected, and Primer Express V2.0 software was used to design probes for Treponema hyodysenteriae and primers. Same as Table 1.
[0051] Primers and probes were synthesized by Shanghai Xuguan Biotechnology Co., Ltd.
Embodiment 2
[0052] Example 2: Extraction of Brevi / Treponema hyodysenteriae DNA
[0053] The DNA of Brevi / Treponema hyodysenteriae was extracted with DNA extraction reagent.
[0054] The specific operation is as follows:
[0055] ① Take n 1.5 mL sterilized centrifuge tubes, where n is the sum of the number of samples to be tested, one tube of positive control and one tube of negative control, and number each tube.
[0056] ② Add 200 μL of DNA Extraction Solution I to each tube, then add 200 μL each of the sample to be tested, the negative control and the positive control, and use a tip for each sample; shake and mix for 5 s on a mixer. Centrifuge at 13000 r / min for 10 min at 4°C~25°C.
[0057] ③ Aspirate and discard the supernatant as much as possible without touching the precipitate, then add 10 μL of DNA Extraction Solution II, shake and mix for 5 s on a mixer. Centrifuge at 2000 r / min for 10 s at 4°C~25°C.
[0058] ④ Dry bath or boiling water bath at 100 ℃ for 10 minutes; centrifu...
Embodiment 3
[0060] Embodiment 3: the establishment of real-time fluorescent PCR
[0061] 1. The concentration of probes and primers, the concentration of magnesium ions and the concentration of enzyme were optimized respectively, and the reaction system of real-time fluorescent PCR was established.
[0062] According to the method of Example 2, the Brevi / Treponema hyodysenteriae DNA was obtained, and the template concentration was about 10 4 -10 6 copy / microliter, perform real-time fluorescent PCR, and optimize the reaction system.
[0063] (1) Optimization of probe and primer concentrations
[0064] Probes and primers were optimized separately. The primer concentration increased by 0.1 μM interval from 0.1 μM to 0.8 μM and the probe concentration increased by 0.1 μM interval from 0.1 μM to 0.5 μM were cross-matched for real-time fluorescent PCR to select the optimal primer and probe concentration. The reaction system conditions are shown in Table 2, and the primers and probe seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com