Kit and method for detecting fish pathogenic bacteria
A pathogenic bacteria and kit technology, applied in the field of kits for detection of fish pathogenic bacteria, can solve the problems of non-representative, non-standard strains, and inability to accurately detect Edwardsiella spp., and achieve strong specificity , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Primer Design Experiment
[0026] 1. Primer design
[0027] Search the conserved gene sequences of Edwardsiella phosphosrine transaminase from GenBank, and design primers based on these sequences. Among them, according to the coding gene sequence of Edwardsiella serC (gene sequence number is AF110153.1, which is a pathogenic gene), a pair of primers designed are:
[0028] Ei1F(5-CATGATAATACCCGGTGTTGG-3)
[0029] Ei1R(5-GTATTGCTGGGGAACAACTC-3),
[0030] As shown in SEQ ID NO.1 and SEQ ID NO.2, the size of the expected amplified fragment is 124bp. Primers were synthesized by Shanghai Sangon Biological Services Co., Ltd.
[0031] 2. Primer verification
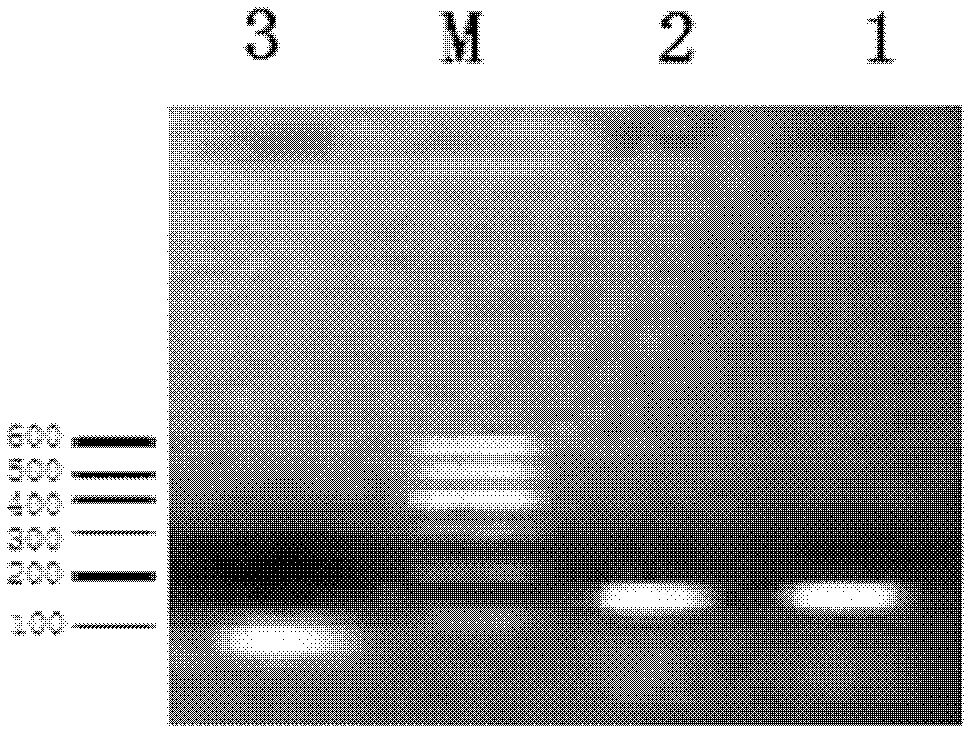
[0032] (1) Template preparation: activating and preparing a bacterial suspension of Edwardsiella catacili ATCC 33202, and extracting genomic DNA. DNA was extracted using Tiangen bacterial genomic DNA extraction kit.
[0033] (2) Amplification: amplify on the Bio-Rad Mycycler gradient amplification instrume...
Embodiment 2
[0040] Specificity and sensitivity detection of embodiment 2PCR detection system
[0041] 1. Experimental method
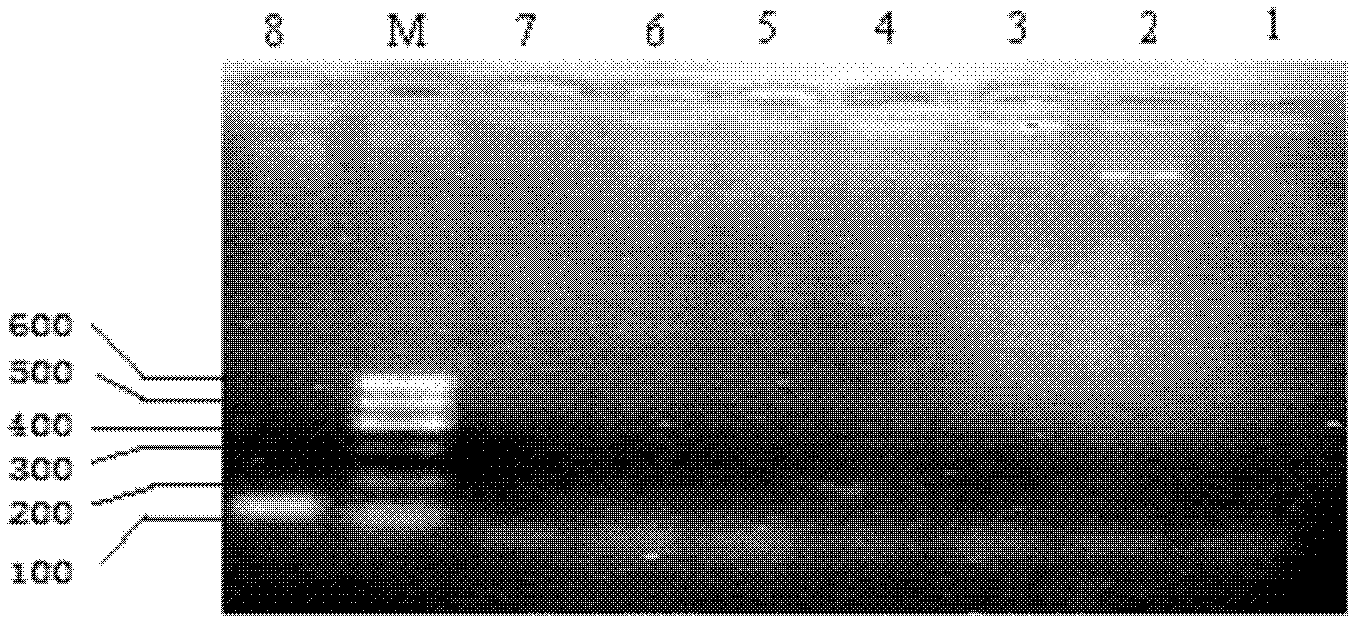
[0042] (1) Template preparation: use the genomic DNA of Flavobacterium columnar, Aeromonas temperatus, Vibrio river, Pseudomonas fluorescens, Aeromonas caviae, Aeromonas hydrophila, and Edwardsiella spp. as templates for determination The specificity of the method; with the bacterium solution of the Edwardsiella spp. (concentration range is 0.28 × 10 8 cfu / mL-0.28×10 1 cfu / mL) and gradient concentrations of genomic DNA (concentration range from 25ng / μL to 2.5fg / μL) are the sensitivity of the template assay method. DNA was extracted using the bacterial genomic DNA extraction kit provided by Tiangen.
[0043] (2) Amplification: amplify on the Bio-Rad Mycycler gradient amplification instrument with the primer pair designed in Example 1;
[0044] 20μL PCR reaction system (final concentration):
[0045]
[0046]
[0047] The PCR program was: pre-denaturation...
Embodiment 3
[0055] Composition and method of use of embodiment 3 test kit
[0056] 1. The composition of the kit
[0057] (1), PCR reaction solution (10 μ L / time): buffer solution, primers with nucleotide sequences as shown in SEQ ID NO: 1-2, Mg 2+ , dNTPs;
[0058] (2), Taq enzyme (0.25μL / time);
[0059] (3), 6% chelex-100 (DNA extraction solution, 200μL / time);
[0060] (4), sterilized ultrapure water.
[0061] 2. How to use
[0062] (1) Extract the genomic DNA of the sample to be tested;
[0063] (2) Using the genomic DNA extracted from the sample to be tested as a template, use the aforementioned kit to amplify:
[0064] 20μL PCR reaction system (final concentration):
[0065]
[0066]
[0067] The PCR program was: pre-denaturation at 95°C for 4 min; denaturation at 94°C for 15 s, annealing at 56°C for 10 s, extension at 72°C for 15 s, and 38 cycles; incubation at 72°C for 5 min and termination at 4°C.
[0068] (3) 2% agarose gel electrophoresis to detect PCR amplificatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com