Epidermal differentiation microRNA signature and uses thereof
An epidermis, expression-level technology, applied in the field of skin, can solve problems such as expensive equipment, inconvenient operation, and multiple follow-up steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
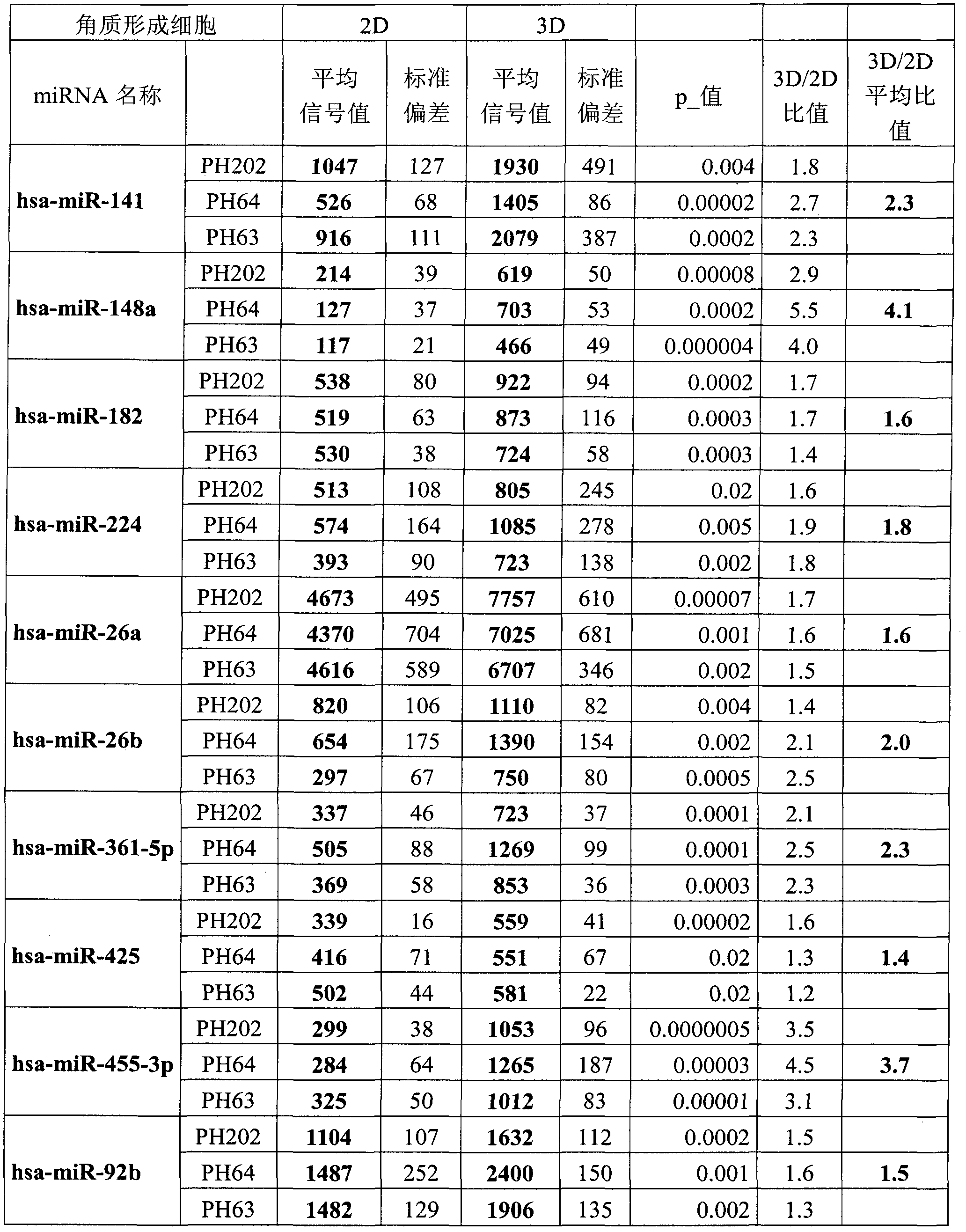
[0196] Example 1: The listed 10 miRNAs are overexpressed in differentiated regenerating epidermal keratinocytes compared to undifferentiated monolayer cultured keratinocytes.
[0197] Three primary normal human keratinocyte cell lines (PH202, PH64, and PH63) derived from mammoplasty were used in this study. The expression of all known human miRNAs was quantified in cultured keratinocyte monolayers, ie undifferentiated keratinocytes (2D) and simultaneously in these same keratinocytes in regenerated epidermal samples (3D).
[0198] The expression of each miRNA was compared statistically between the "keratinocytes in culture" sample and the "regenerated epidermal keratinocytes" sample, and compared using a Student's t-test giving the probability p. For each cell line studied, the expression level of each miRNA listed in Table 1 in the epidermis was statistically significantly higher than that in undifferentiated keratinocytes (p<0.05). For each cell line, the expression ratio 3D...
example 2
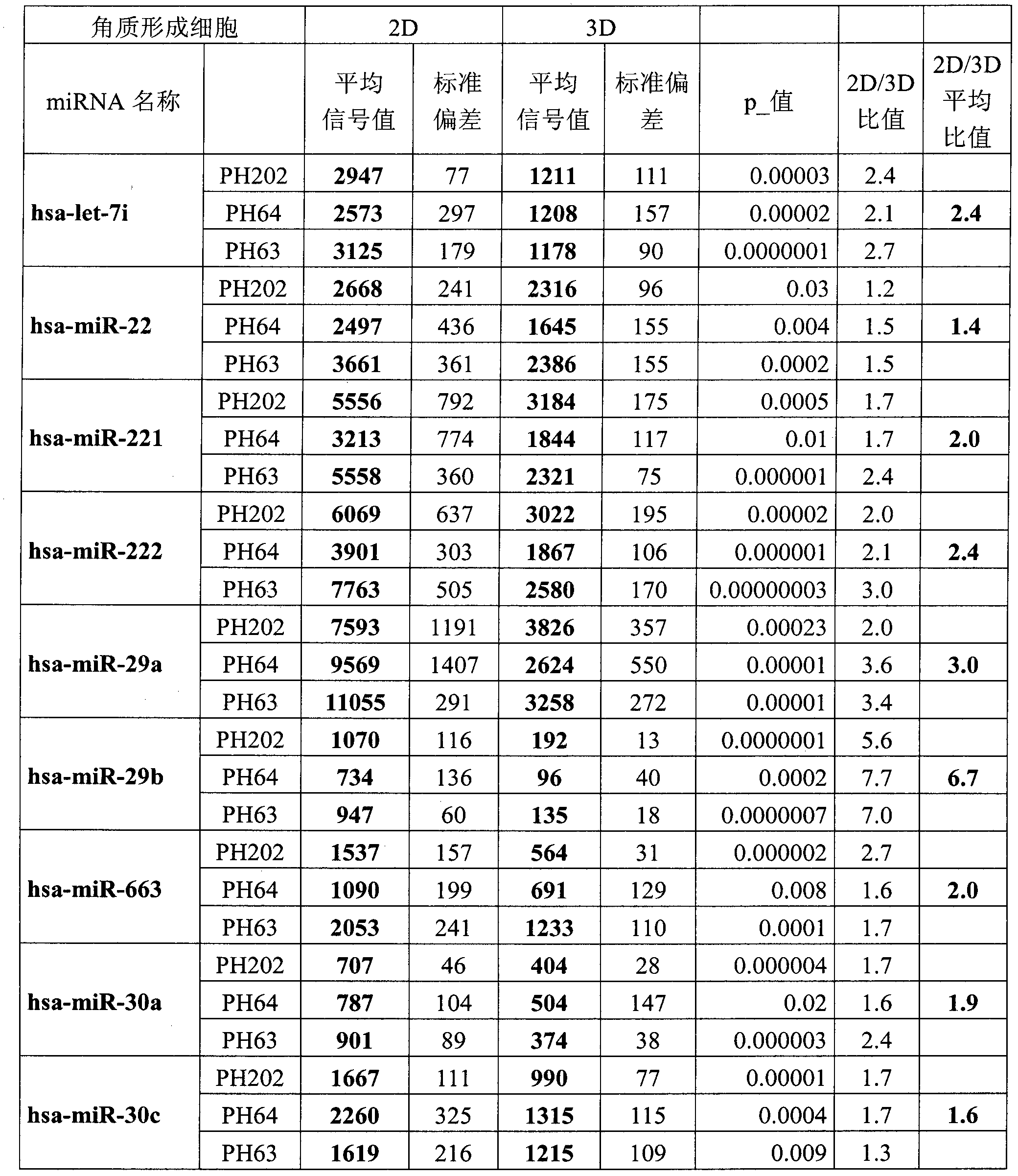
[0201] Example 2: Nine miRNAs listed are overexpressed in undifferentiated monolayer cultured keratinocytes compared to differentiated regenerating epidermal keratinocytes.
[0202]Three primary normal human keratinocyte cell lines (PH202, PH64, and PH63) derived from mammoplasty were used in this study. The expression of all known human miRNAs was quantified in cultured keratinocyte monolayers, ie undifferentiated keratinocytes (2D) and simultaneously in these same keratinocytes in regenerated epidermal samples (3D). The expression of each miRNA was statistically compared between the "cultured keratinocytes" sample and the "regenerated epidermal keratinocytes" sample, and compared using a Student's t-test for which probability p was obtained. For each cell line studied, the expression levels of each miRNA listed in Table 1 were statistically higher in the epidermis than in undifferentiated keratinocytes (p<0.05). The expression ratio 3D / 2D is given for each cell line. For e...
example 3
[0205] Example 3: Method for detecting miRNAs.
[0206] Northern Blot:
[0207] Qualitative and quantitative using conventional methods. Multiple miRNAs can be analyzed in the same experiment. This method requires the use of 5-50 μg of total RNA.
[0208] RT Q-PCR
[0209] This method can study the precursors of miRNAs and is suitable for studying mature miRNAs. This method is commercially available (TaqMan MicroRNA Assays from Applied Biosystems) and is described in several articles [23].
[0210] miRNA Microarray Analysis (Microarray)
[0211] This method is similar to conventional mRNA microarray analysis, and is based on fluorescent labeling of complementary cDNAs of miRNAs in samples, and immobilizing these labeled cDNAs, hybridizing on slides loaded with immobilized miRNA sequences [25].
[0212] Some authors argue that this miRNA microarray technology is an option for profiling the global expression profile of miRNAs if the novel sequences of miRNAs derived ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com