Method for detecting Enterobacter sakazakii by employing non-labeled fluorescence PCR technology and HRM analysis technology
A technology of Enterobacter sakazakii and labeled fluorescence, which is applied in the field of microbial detection and can solve the problems of complex operation, high detection cost, and cumbersome operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
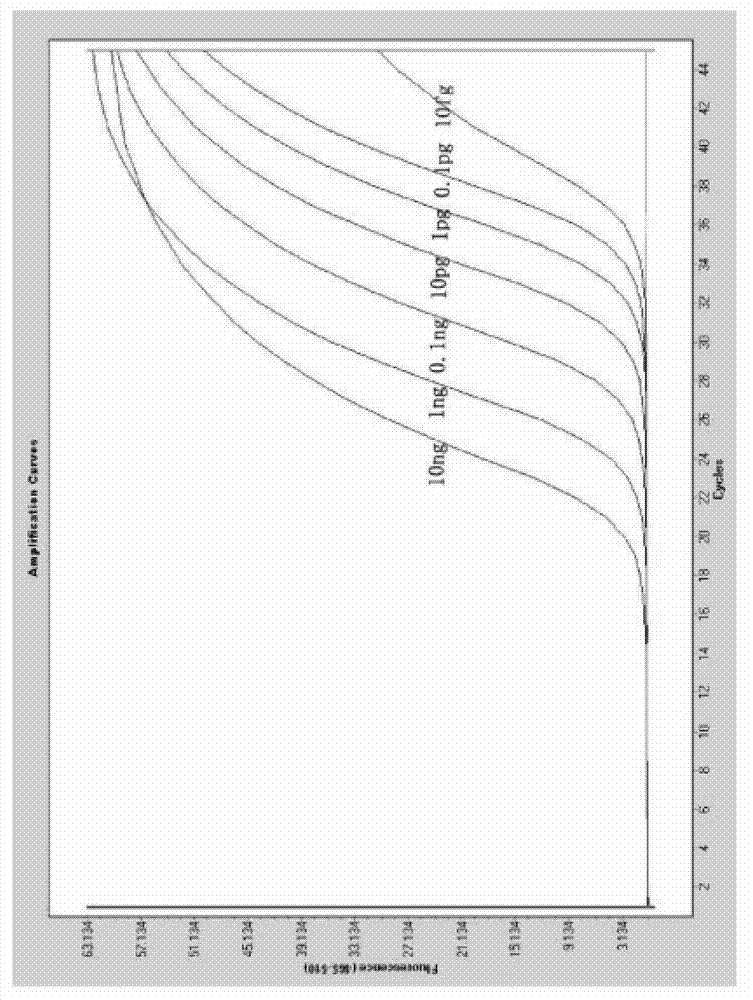
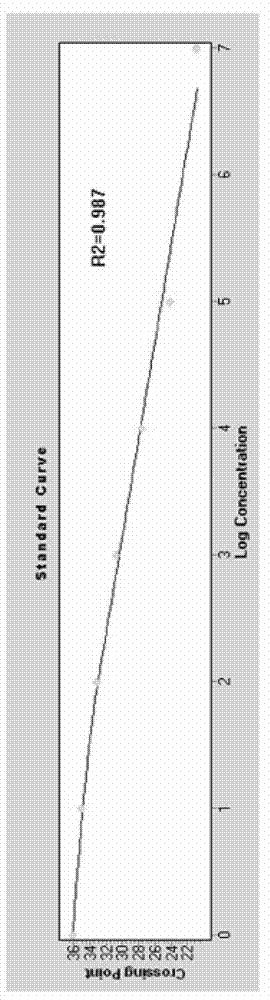
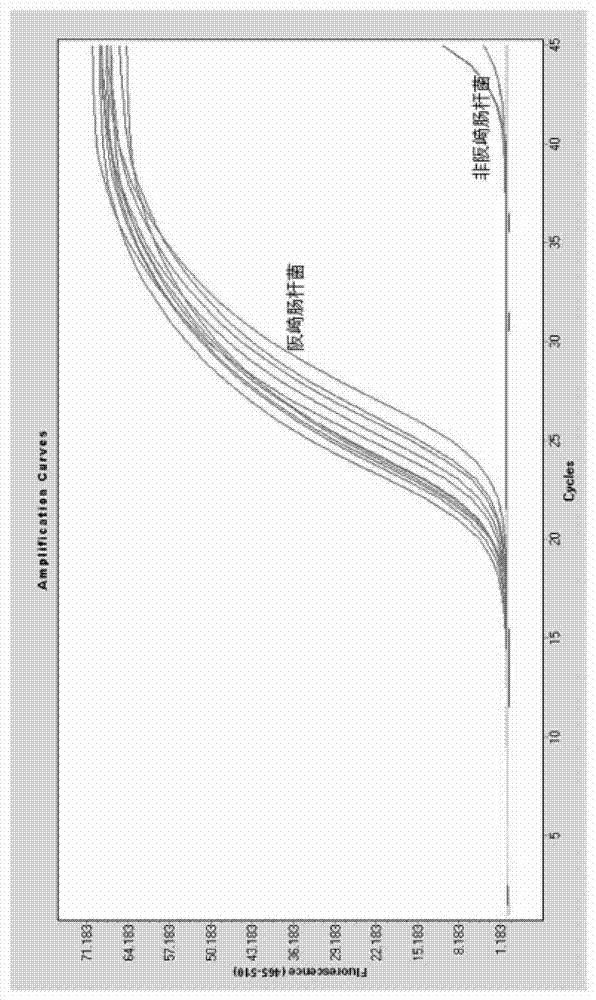
[0064] Detection of Enterobacter sakazakii in milk powder using non-labeled fluorescent PCR combined with HRM analysis technology of the present invention
[0065] DNA extraction
[0066] Weigh 25 g of the sample by aseptic operation, add it to 225 mL of nutrient broth for cultivation, and incubate at 36±1°C for 18 hours. Take 1.5 mL of the culture and centrifuge at 10,000 rpm for 2 min; add 500 μL of TE buffer to the precipitate, repeatedly pipette to resuspend it, add 30 μL of 10% SDS and 15 μL of proteinase K, mix well, and incubate at 37°C for 1 hour; NaCl, mix well, then add 80ul CTAB / NaCl solution, mix well and then incubate at 65°C for 10min; add an equal volume of phenol / chloroform / isoamyl alcohol and mix well, centrifuge for 4-5min, transfer the supernatant to a In a new tube, add 0.6-0.8 times the volume of isopropanol, mix gently until the DNA precipitates, and the precipitate can be centrifuged slightly; after the precipitate is washed with 1mL of 70% ethanol, cen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com