Real-time fluorescence quantification PCR (Polymerase Chain Reaction) kit and method for detecting Y-chromosome micro-deletion
A real-time fluorescence and Y-chromosome technology, applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of low detection throughput, high cost, inapplicability, etc., to reduce time and The effect of cost, high sensitivity and specificity, and high detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
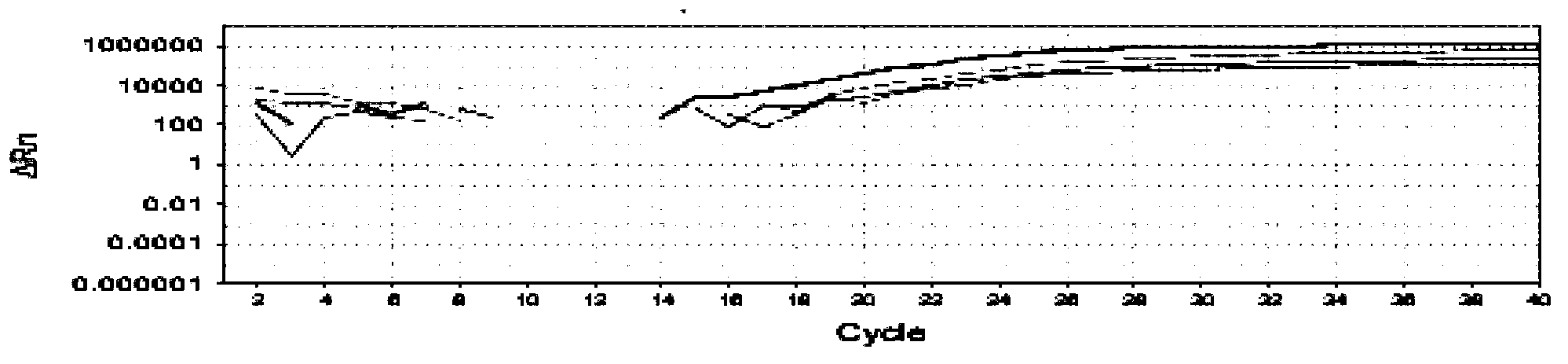
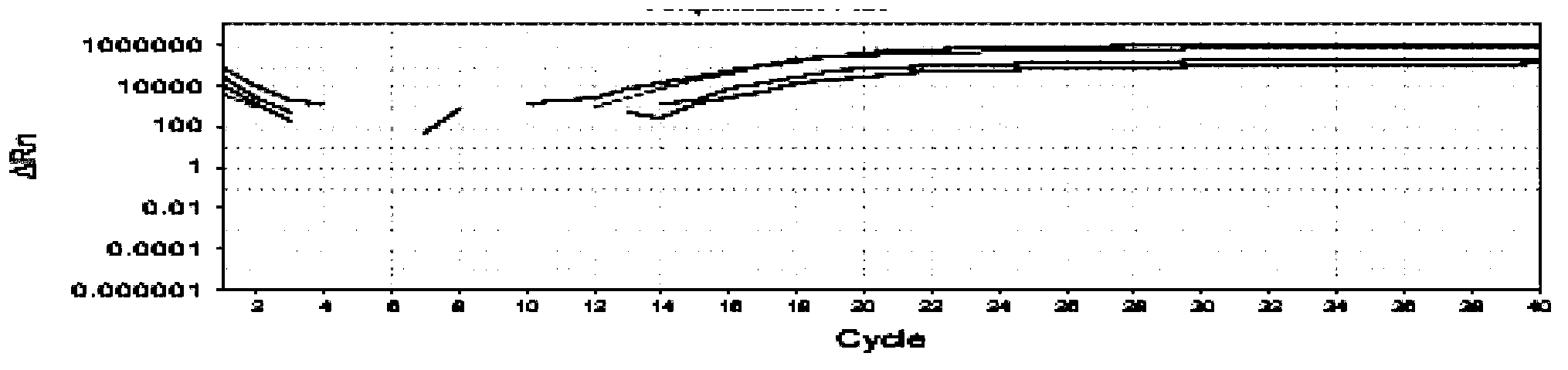
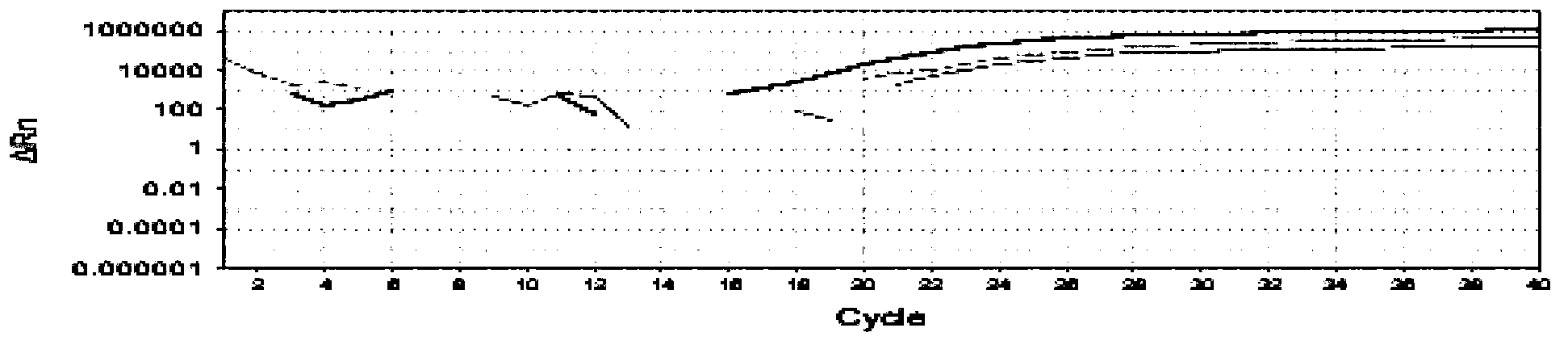
Image
Examples
Embodiment 1
[0066] 1. Materials
[0067] 1. Instrument: Real-time Fluorescence Quantitative Instrument
[0068] Design of probes and primers: 6 pairs of probes and primers were designed to specifically detect AZF sequences based on the above, and at the same time, 1 pair of probes and primers were designed as quality control to detect male sex-determining genes to detect whether the PCR detection process is normal To carry out, the normal fertile male DNA sample is used as a positive control, the female DNA sample is used as a negative control, and sterilized water is used as a blank control to prevent contamination of the test system.
[0069] 2. Use the following probe primer pairs to detect the deletion of AZFa, AZFb, and AZFc regions.
[0070] SY84 upstream primer 5'-CCTATTTGTTTTAAGGTGCCATTCC-3' (SEQ ID NO: 1)
[0071] SY84 downstream primer 5'-GCTTGCCTGAGCATCCTACAG-3' (SEQ ID NO: 9)
[0072] SY84 probe 5'-VIC-CTCTACCTCCTTCCCCCAGTGCCCA-BHQ-3' (SEQ ID NO: 15)
[0073] SY86 upstream...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com