SiRNA interfering GDF9 gene expression and application thereof
A technology for gene expression and expression vectors, applied in the fields of molecular biology and bioengineering, to achieve good silencing effects and reduced protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Construction of pCDNA3.1-GDF9 eukaryotic expression vector
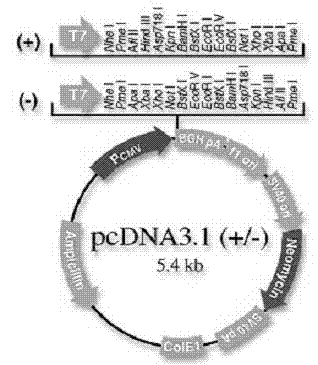
[0038] 1. Synthesize the buffalo growth and differentiation factor 9 (GDF9) sequence (GenBank accession number NM_174681), connect it to the vector pUC19, and cut it into Eco RI and BamH. double digestion ( Eco R I and BamH) and sequencing proved that the synthesized fragments were correct (see figure 2 ).
[0039] 2. Construction of pCDNA3.1-GDF9 eukaryotic expression plasmid
[0040] (1) Enzyme digestion reaction
[0041] reaction system: Eco R I and BamHI double-digest the PCR product. Eco R I and BamHI double-digest pcDNA3.1(-), enzyme digestion system: 10×Buffer K 2 μL, Eco R I 1 μL, BamHI 1 μL, plasmid 6 μL, distilled water 10 μL, total volume 20 μL. Reaction conditions: act at 37°C for 1.2h, and perform 1% agarose gel electrophoresis. The digested products were purified with a DNA purification kit, and finally the DNA was eluted with 30 μL of water and stored at -20°C.
[004...
Embodiment 2
[0046] Example 2 Construction of pshRNA-copGFP Lentivector lentiviral interference plasmid
[0047] 1. According to the method and principle of buffalo growth and differentiation factor 9 mRNA sequence design, target sequences were screened out, and siRNA was designed to reversely complement buffalo growth and differentiation factor 9 gene mRNA, which were named GDF9-1, GDF9-2, and GDF9-3, respectively. Its sense strand sequence is as follows:
[0048] GDF9-1: SEQ ID NO: 1;
[0049] GDF9-2: SEQ ID NO: 2;
[0050] GDF9-3: SEQ ID NO:3.
[0051] 2. According to the siRNA target sequence, design the DNA sequence required to construct the siRNA vector that inhibits the expression of buffalo growth differentiation factor 9 gene: GDF9 shRNA template, introduced at both ends Bam H I and Eco The R I enzyme cutting site was sent to Shanghai Bioengineering Technology Service Co., Ltd. for synthesis. The specific sequence is as follows (F indicates the sense strand, R indicates th...
Embodiment 3
[0064] Example 3 Screening of Effective Targets of RNA Interference
[0065] 1. Transfection of eukaryotic cells with three lentiviral interference plasmids
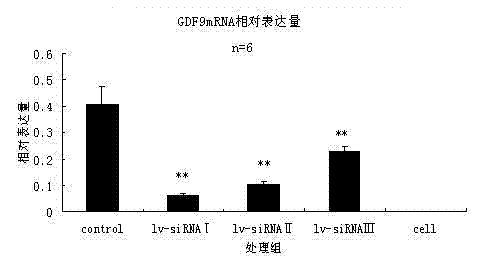
[0066] pcDNA3.1-GDF9 and LV-siRNAⅠ, LV-siRNAⅡ and LV-siRNAⅢ were co-transfected into CHO cells for expression. CHO cells were co-transfected with pcDNA3.1-GDF9 and LV-shRNA-GFP empty plasmids as a control, in order to be consistent with the transfection background of the treatment group, and 6 replicates were set for each treatment. The ratio of pcDNA3.1-GDF9 to lentiviral interference plasmid is 1:1.
[0067] Lipofection was carried out according to the instruction manual of Polyfect (purchased from QIAGEN).
[0068] (1) One day before transfection, inoculate cells in a new six-well culture dish, and completely blow off the cells to ensure that the cells are evenly distributed. When transfecting, the cells should ideally cover 40-90% of the dish.
[0069] (2) Add pcDNA3.1-GDF9 and lentivirus interference plasmid a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com