Method for detecting fungal pathogens
A pathogenic, fungal technology used in the detection of fungal pathogens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment -1
[0100] Cultivation of the fungal pathogens used in this study: The following are the growth conditions used for the cultivation of the fungal pathogens of apples.
[0101] Alternaria alternata
[0102] Growth medium - PDA (Potato Dextrose Agar; Himedia); Growth conditions - Aerobic
[0103] Temperature: 28°C; Culture time: 5 days
[0104] Glomerella cingulata
[0105]Growth medium - PDA (Potato Dextrose Agar; Himedia); Growth conditions - Aerobic
[0106] Temperature: 25°C; Culture time: 5 days
[0107] Colletotrichum acutatum
[0108] Growth medium - PDA (Potato Dextrose Agar; Himedia); Growth conditions - Aerobic
[0109] Temperature: 25°C; Culture time: 5 days
[0110] Botrytis cinerea
[0111] Growth medium - PDA (Potato Dextrose Agar; Himedia); Growth conditions - Aerobic
[0112] Temperature: 25°C; Culture time: 5 days
[0113] Apple scab (Venturia inaequalis)
[0114] Growth medium-M-98 (wort juice 10gm / L; peptone 3gm / L and agar 15gm / L; Himedia); growth condit...
Embodiment -2
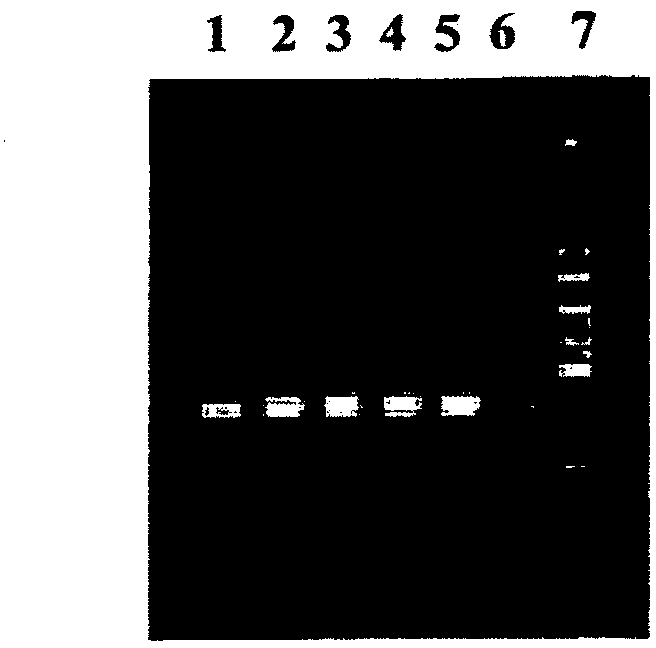
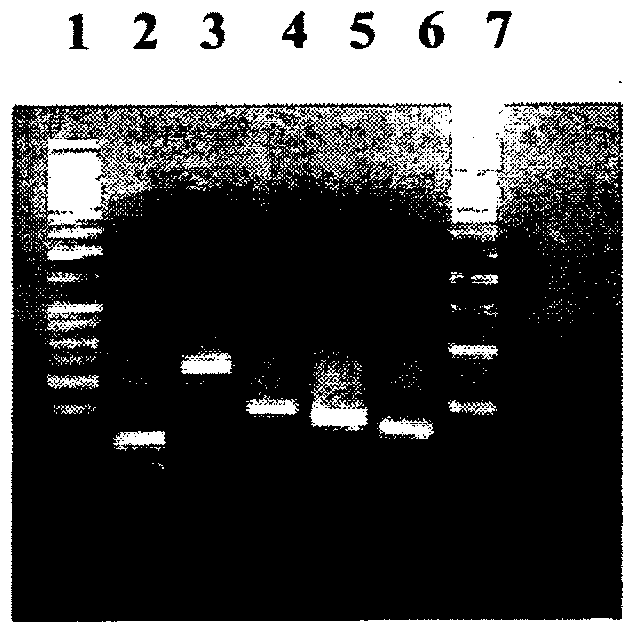
[0116] Design of universal fungal-specific primers: rDNA sequences of various apple fungal pathogens were downloaded from NCBI (http: / / www.ncbi.nlm.nih.gov). In order to obtain the conserved regions in these sequences, multiple sequence alignments were performed using CLUSTALW2 (http: / / www.ebi.ac.uk / Tools / clustalw2 / index.html). Both the forward primer (ITRRF1: 5'CGATGAAGAACGCAGCGAAAT) and the reverse primer (ITRRRI: TATGCTTAAGTTCAGCGGGTATC) were designed from the conserved region flanking the ITS2 region of the fungal rDNA sequence.
Embodiment -3
[0118] Analysis of the generality of the designed primers: To find out whether the primers designed in this study could amplify the rDNA sequences of other fungi, we downloaded the rDNA sequences of a variety of randomly selected fungi [Debaryomyces hansenii Strain W4682 (GQ913348); Fungal endophyte isolate 5724 (DQ979775); Cladosporium cladosporioides strain STE-U (AY251074); Cladosporium cladosporioides isolate GG6 ( EF 104249); Athelia bombacina isolate AFTOL-ID347 (DQ449026); Chaetomium globosum isolate B221 (GQ365152); Microsphaeropsis sp. HLS303 (FJ770074; Microsphaeropsis arundinis (Microsphaeropsis arundinis) strain PMBMDF049 (FJ798603); Dothideomycetes sp. 11313 (GQ153229); Aspergillus flavus strain ATCC11497 (GU256760); Aspergillus terreus strain ATCC2056475 (9GU2); Aspergillus niger strain ATCC64973 (GU256750); Aspergillus versicolor strain ATCC26644 (GU256744); Debaryomyces hansenii strain W4682 (GQ913348); Neurospora crassa ) strain ATCC MYA-4619 (GU327635); Neur...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com