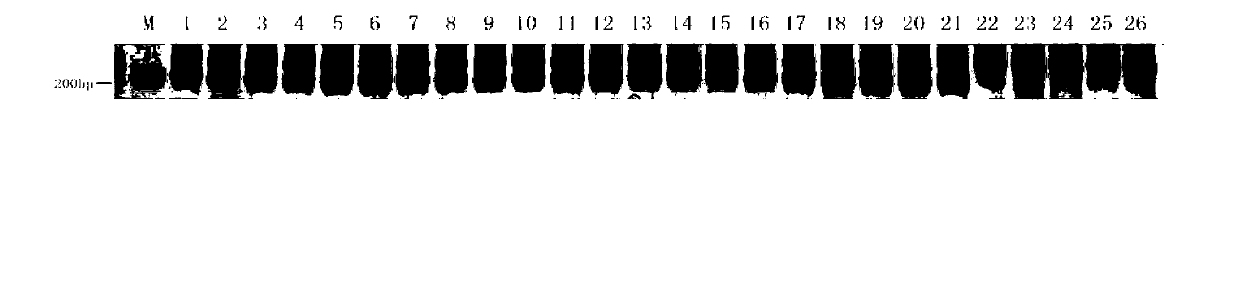Rapid detection method for purity of cucumber seeds
A detection method and seed technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of human factors and environmental factors, low accuracy rate, laborious and other problems, and achieve manpower saving and high accuracy , determine the effect of simple
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: (Using three pairs of primers to make the standard map of cucumber variety 'Zhexiu No. 1' respectively)
[0032] (1) Extraction of DNA: Take 3 seeds of 'Zhexiu No. 1', remove the seed coat, and extract genomic DNA by CTAB method;
[0033] (2) PCR amplification: The total PCR amplification reaction system is 20μl, and the amplification reaction system components are: Zhexiu No. 1'seed genomic DNA 20ng each, 10pmol / μl primers CSⅥ-37.2-F770, CSⅥ-24.9-F757 and CSⅤ-18.5-F664 upstream primer, downstream primer each 0.4μl, 2.5mM Mg 2+ 2μl, 2mM dNTP 1μl, 5U / μl Taq DNA polymerase 0.1μl, 10×buffer 2μl, sterile distilled water to make up to 20μl;
[0034]The amplification program of the PCR reaction is: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 sec, annealing at 55°C for 1 min, extension at 72°C for 1 min, 35 cycles, extension at 72°C for 10 min, and storage at 4°C;
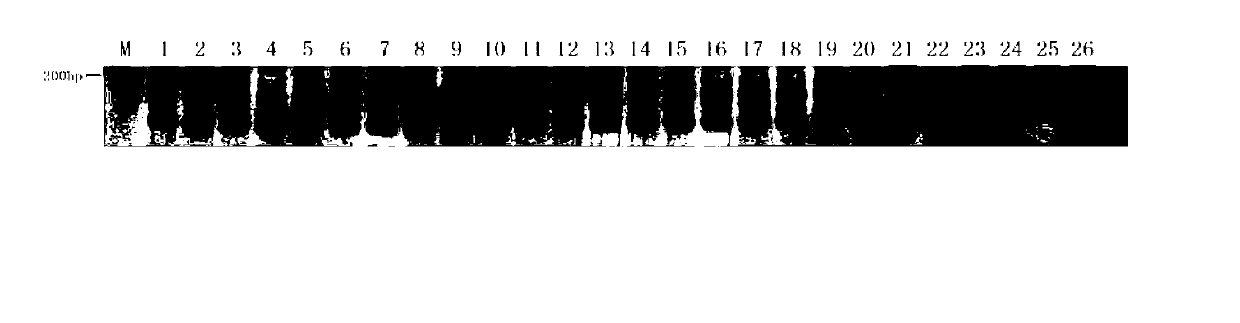
[0035] (3) Detection of amplified products and preparation of the standard m...
Embodiment 2
[0038] Example 2: (Detection of 'Zhexiu 1' and its parents and mixed seeds using primer CSⅥ-37.2-F770)
[0039] (1) DNA extraction: Take 20 seeds of 'Zhexiu 1', 1 seed of the male parent 'XD23', 1 seed of the female parent 'XH65' and 4 seeds of other cucumber inbred lines artificially mixed in, after removing the seed coat, Genomic DNA was extracted by CTAB method;
[0040] (2) PCR amplification: The total PCR amplification reaction system is 20 μl, and the amplification reaction system components are: Zhexiu No. 1', male parent, female parent, and samples to be tested (in this example, other cucumber inbred lines artificially mixed 20ng of seed genomic DNA, 10pmol / μl of primer CSⅥ-37.2-F770, 0.4μl of upstream primer and downstream primer of primer CSⅥ-37.2-F770, 2.5mM Mg 2+ 2μl, 2mM dNTP 1μl, 5U / μl Taq DNA polymerase 0.1μl, 10×buffer 2μl, sterile distilled water to make up to 20μl;
[0041] The amplification program of the PCR reaction is: pre-denaturation at 94°C for 5 ...
Embodiment 3
[0045] Example 3: (Rapid detection of 'Zhexiu No. 1', parents and samples to be tested using primer CSⅥ-24.9-F757)
[0046] (1) Extraction of DNA: Take 20 seeds of 'Zhexiu 1', 1 seed of the male parent 'XD23', 1 seed of the female parent 'XH65' and 4 seeds of the male parent 'XD23' (in this case, artificially mixed male parent seeds As the cucumber seeds to be tested), after removing the seed coat, the genomic DNA was extracted by the CTAB method;
[0047] (2) PCR amplification: The total PCR amplification reaction system is 20μl, and the amplification reaction system components are: Zhexiu No. 1', 20ng each of male parent, female parent and sample to be tested, 10pmol / μl primer CSⅥ -24.9-F757 upstream primer, downstream primer 0.4μl, 2.5mM Mg 2+ 2μl, 2mM dNTP 1μl, 5U / μl Taq DNA polymerase 0.1μl, 10×buffer 2μl, sterile distilled water to make up to 20μl;
[0048] The amplification program of the PCR reaction is: pre-denaturation at 94°C for 5 min, denaturation at 94°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com