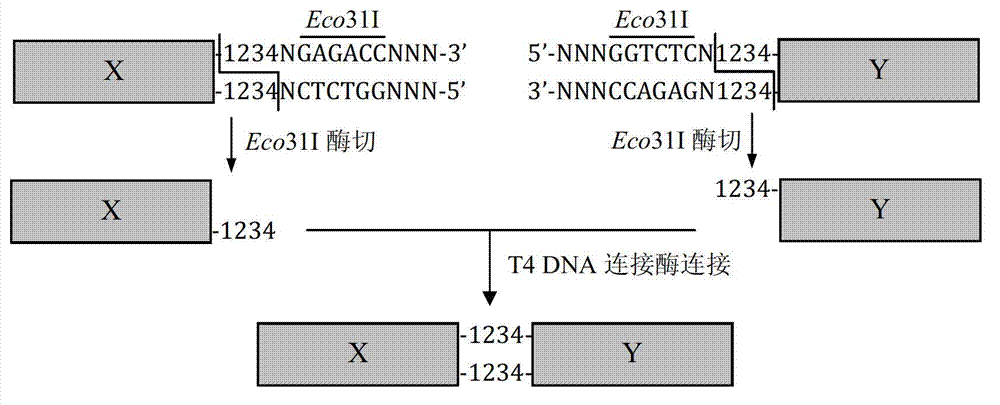
DNA (Deoxyribonucleic Acid) multi-site directed mutation method
A directional mutation and multi-site technology, applied in the field of genetic engineering, can solve the problems of not widely used, increased workload, time-consuming and labor-intensive, etc., and achieve the effects of wide application range, improved efficiency and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 Aspergillus niger salicylate hydroxylase Aspergillus niger salicylate hydroxylase (AnSh, Genbanknumber: NT_166526) gene intron removal
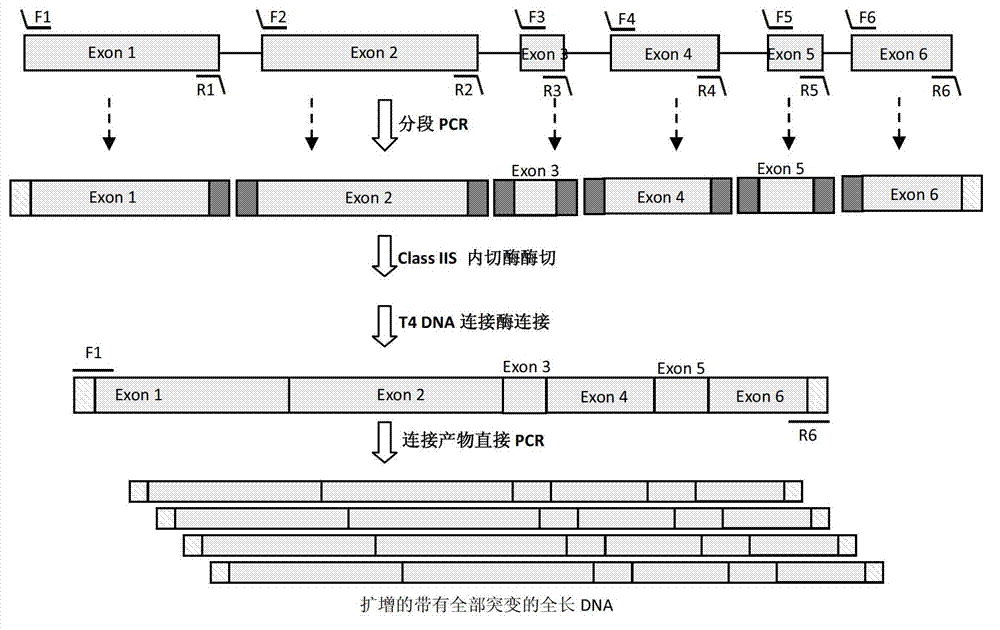
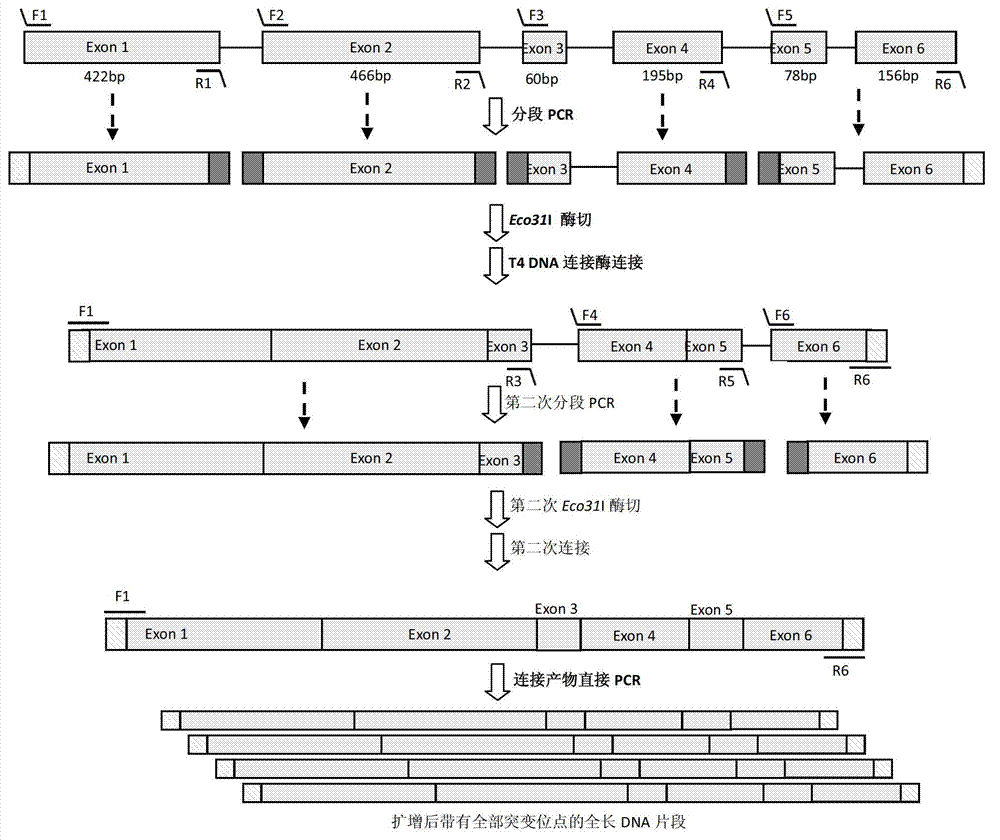
[0054] The full-length gene of Aspergillus niger salicylate hydroxylase (AnSh, Genbank number: NT_166526) is 1718bp, including 5 introns and 6 exons, such as image 3 shown. image 3 In , the long boxes represent exons, and the lines represent introns. The short lines above and below the long box represent primers used for PCR amplification, wherein the short lines above represent forward primers, for example, F1, F2, F3, F4, F5, F6. The dashes below indicate reverse primers, eg, R1, R2, R3, R4, R5, R6. Among them, the external primers are F1 and R6. Internal primers are F2, F3, F4, F5, F6, R1, R2, R3, R4, R5.
[0055] image 3 In the figure, the small squares on both sides of the long box represent the restriction endonuclease sites introduced after segmental PCR amplification, and the dark squares represent Class IIS r...
Embodiment 2
[0090] Example 2 Repair of Aspergillus niger salicylate hydroxylase base deletion mutant
[0091] In this example, the base deletion mutant of Aspergillus niger salicylate hydroxylase was repaired. A mutant of Aspergillus niger salicylate hydroxylase has 4 base deletion mutations at 888-890bp and 894bp, such as Figure 5 Sequence comparison of the four base deletion mutations of the AnSh gene shown. Figure 5 In , the base position where the mutation occurs is indicated in the box. The bases marked in italics and underlined are the target gene regions referred to in primer design. Among them, the base CACC in black font is the complementary sequence of the proposed junction.
[0092] In this embodiment, the primers designed according to the DNA mutation site are as follows:
[0093] External primer: the same as 1F (forward primer) and R6 (reverse primer) in Example 1.
[0094] Internal primers:
[0095] P1R: 5′ATA T CACCCAGAGACGCCA 3′;
[0096] P2F: 5′GGC G AACC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com