Efficient splitting tandem gene, efficient splitting plasmid and construction method and appliance
A technology for tandem genes and cleavage genes, applied in the field of efficient lysis plasmids and construction methods for effectively cleaving tandem genes, to achieve the effect of reducing lateral transmission and increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
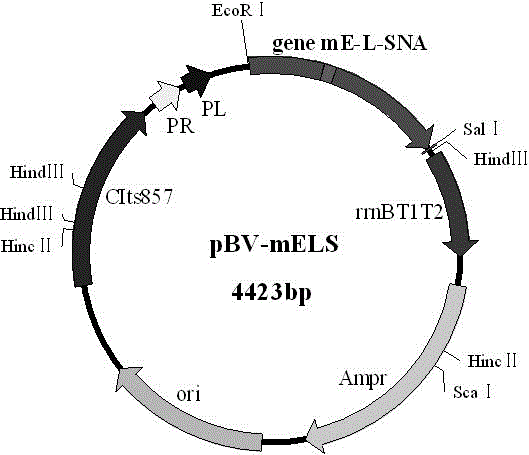
[0040] Example 1, Construction of Safe and Efficient Lysis Plasmid pBV-mELS
[0041] 1.1 PCR amplification of mutant cleavage gene E of phage PhiX174
[0042] Primers were designed according to the coding sequence of PhiX174 (GenBank No. J02482.1) cleavage gene E in GenBank, and the mutant cleavage gene E (mE) was amplified by introducing point mutations into the primers. Introduced at the 5' end of primer mE-F Eco R I restriction enzyme site, 15 amino acids (Gly) were introduced at the 5' end of primer mE-R 4 Ser) 3 The sequence is used as a Linker, and the length of the amplified fragment is expected to be 327 bp. The primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd., and the sequences are as follows:
[0043] mE-F: 5'-GC GAATTC TGGTACGCTGGACTTTGTG-3', (see sequence 4 in the sequence listing)
[0044] mE-R: 5'-GC AGAACCACCACCACCAGAACCACCACCACCAGAACCACCACCACC CTCCTTCCGCAC-3', (see sequence 5 in the sequence listing)
[0045] The mutant cleavage...
Embodiment 2
[0055] Embodiment 2, the preparation of safe Escherichia coli DH5α slough
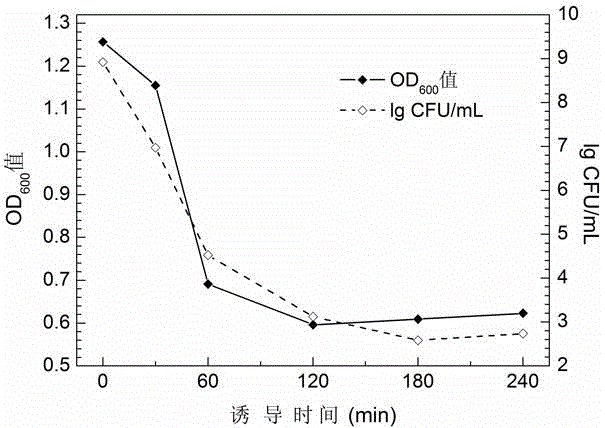
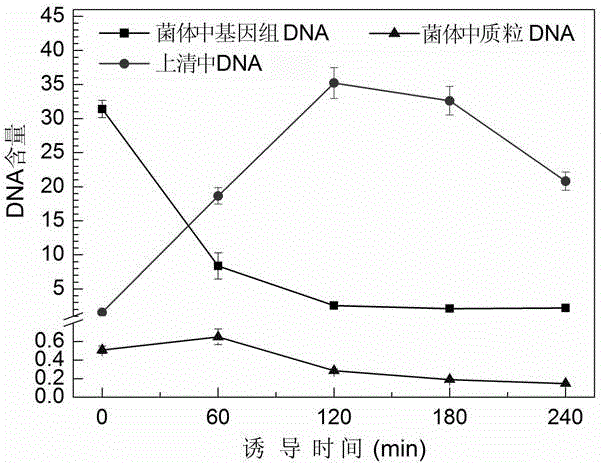
[0056] 2.1 Induced expression of highly efficient cleavage tandem gene mE-L-SNA
[0057] Inoculate Escherichia coli DH5α (pBV-mELS) monoclonals transformed with high-efficiency cleavage plasmids into 5 mL LB medium containing 100 μg / mL ampicillin, culture with shaking at 30°C overnight, and then transfer to 50 mL In LB medium containing 100 μg / mL ampicillin, shake culture at 30°C until OD 600 When the value reached 1.0-1.2, the bacterial culture was rapidly shifted to 42°C to induce the expression of the tandem gene mE-L-SNA. In order to maximize the activity of SNA, a final concentration of 10 mM CaCl was added at 90 min of warming induction 2 and 1 mM MgCl 2 . Samples were taken at intervals before and after induction to measure the OD of the bacterial solution 600 Value and viable count to monitor bacterial growth and lysis. After the end of the induction, the slough formed was washed with P...
Embodiment 3
[0065] Embodiment 3, the lytic activity comparison of plasmid pBV-mELS and pBV-mE
[0066] 3.1 Construction of lysis plasmid pBV-mE
[0067] Apply primers mE-F and mE-R' (5'-CT GTC GAC TCACTCCTTCCGCACGTA -3') (introduced at the 5' end Sal Ⅰ restriction enzyme cutting site), using PhiX174 RFI DNA as a template to amplify the mutant cleavage gene E by PCR, and apply it Eco R I and Sal Insert the plasmid pBV220 after double digestion with I restriction endonuclease, and then transform Escherichia coli DH5α competent cells. Positive clones identified by enzyme digestion were then identified by sequencing. The plasmid with the correct sequence is the pBV-mE plasmid.
[0068] 3.2 Comparison of cleavage kinetics of plasmids pBV-mELS and pBV-mE
[0069] The method of inducing lysis of Escherichia coli DH5α (pBV-mE) is basically the same as that of Escherichia coli DH5α (pBV-mELS), the only difference is that Escherichia coli DH5α (pBV-mE) does not need to add CaCl during ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com