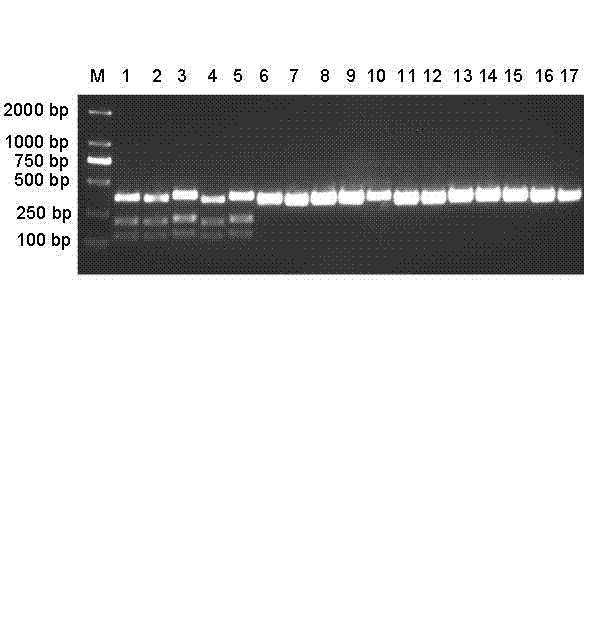
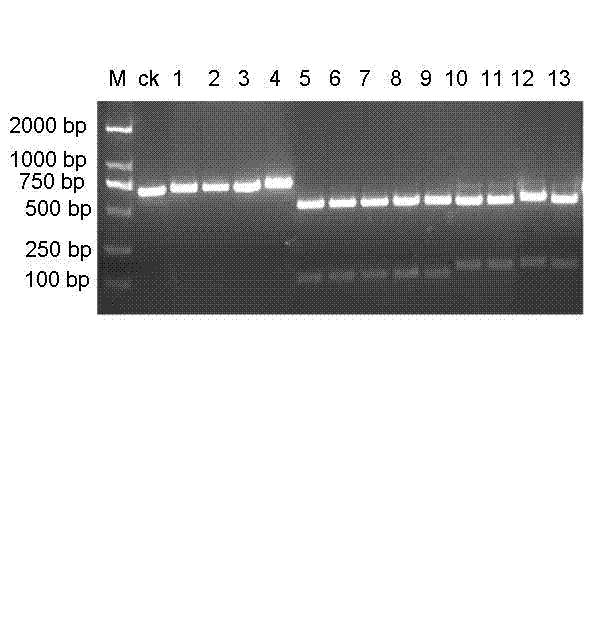
PCR-RFLP (Polymerase chain reaction-restriction fragment length polymorphism) method for quickly identifying panax notoginseng and analogue panax stipuleanatus
A technology of Panax notoginseng and Wenshan, which is applied in the field of molecular identification of species, can solve the problems of sensory identification of the authenticity of Wenshan Sanqi raw materials, easy to mix into the screen side Sanqi, etc., and achieve the effect of overcoming the authenticity problem and accurate identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0015] Main instruments of the present invention: PCR instrument (CFX Manager), Hitachi desktop centrifuge, nucleic acid and protein analyzer (GeneQuant pro), water bath, refrigerator, electrophoresis instrument (DYY-6C), gel imaging analysis system (ImageQuant300). The following operations are conventional methods unless otherwise specified.
[0016] 1. Extraction of sample DNA
[0017] Wenshan notoginseng leaves and roots are collected from three-year-old plants in the Sanqi Science and Technology Demonstration Garden in Miao Township, Wenshan Zhuang and Miao Autonomous Prefecture, Yunnan Province; Pingbian notoginseng leaves and roots are collected from Wenshan notoginseng, Gulinqing Township, Maguan County, Wenshan Zhuang and Miao Autonomous Prefecture, Yunnan Province In the science and technology demonstration park, the leaves and roots of Wenshan notoginseng and Pingbian notoginseng were taken respectively, and DNA was extracted respectively. DNA was extracted accordin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com