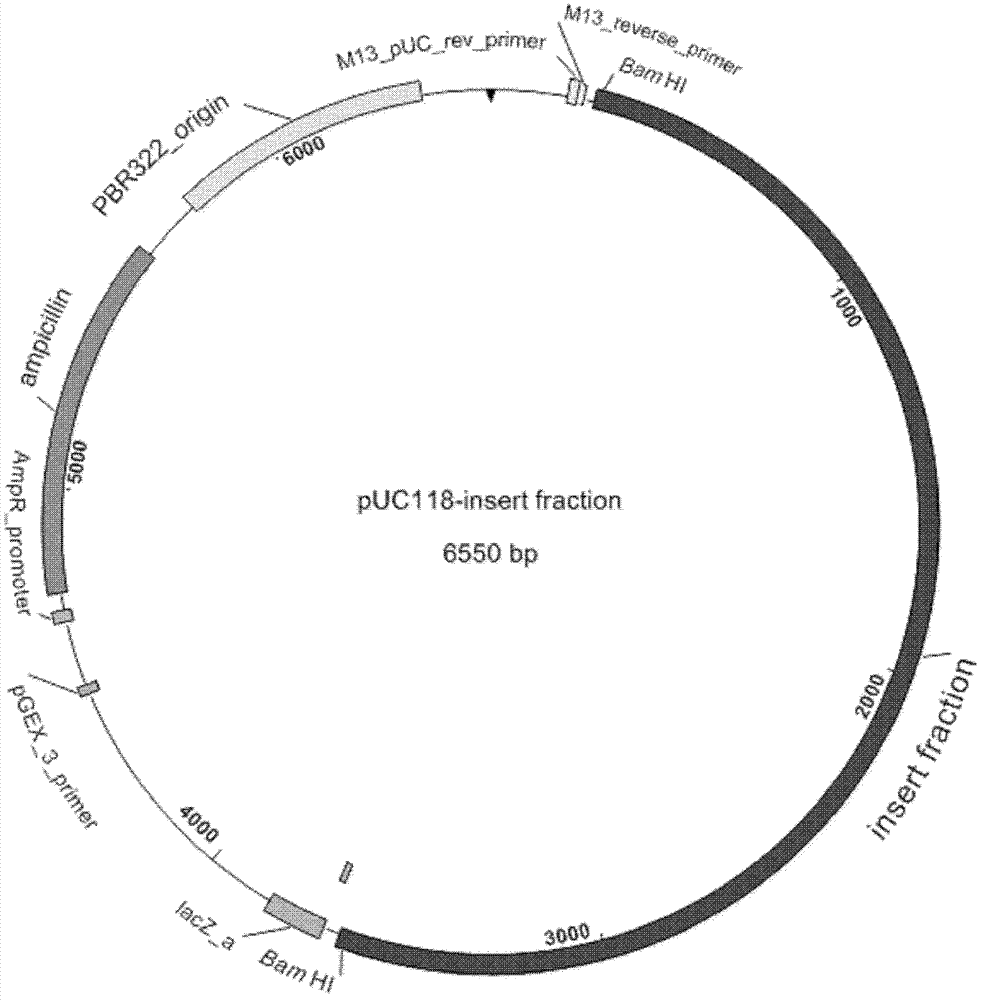
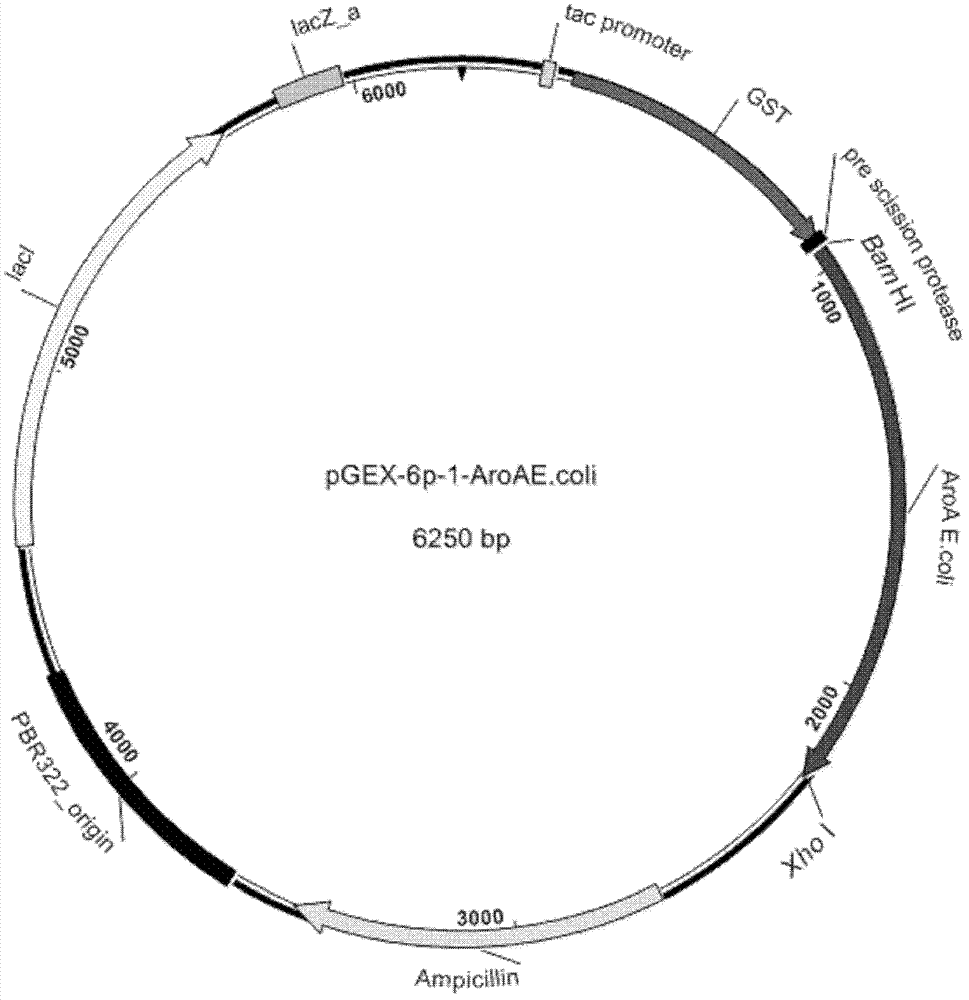
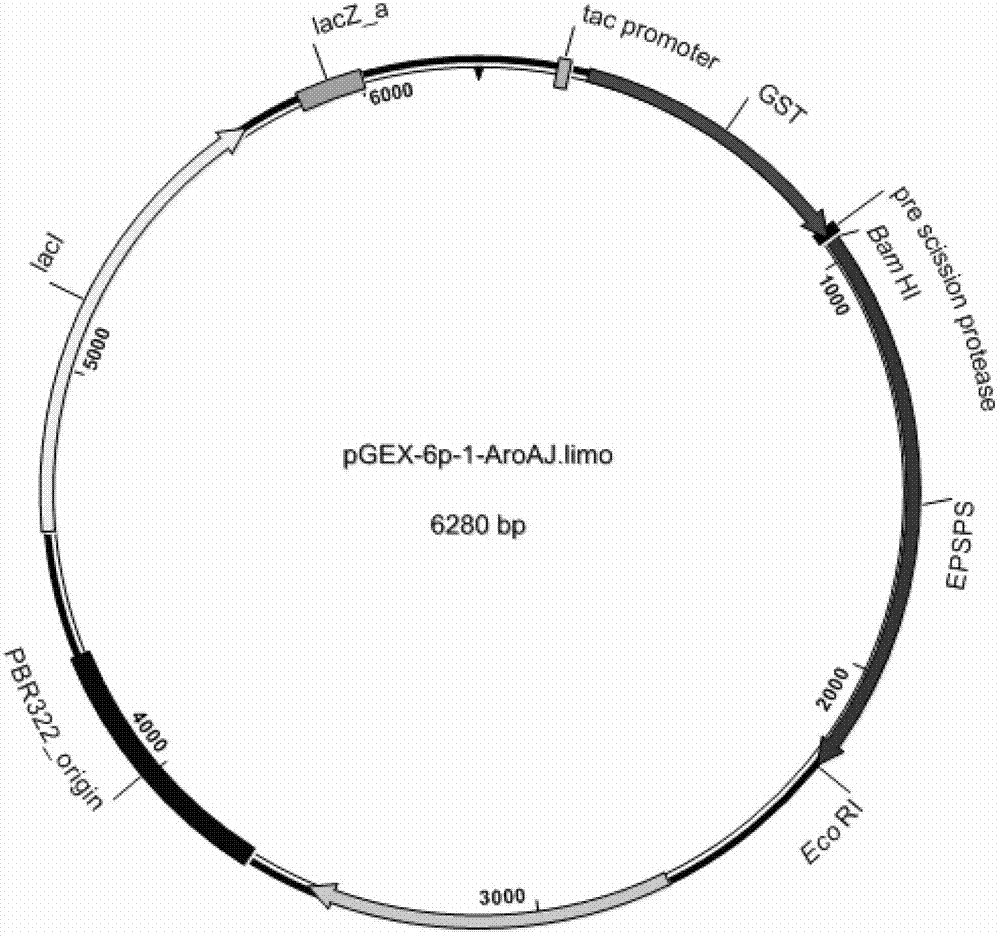
Isolation of 5-enol acetone shikimic acid-3-phosphate synthase gene
A technology of enolacetone shikiki and phosphate synthase, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Isolation and cloning of 5-enolpyruvylshikimate-3-phosphate synthase gene
[0032] (1) Construction and sequencing of genomic library to obtain resistance genes
[0033] The original Janibacter limosus 1C00094 strain (also known as Janibacter limosus 1C00094) with glyphosate resistance of 150mM was selected from a natural deep-sea marine bacterium screened by the Third Research Institute of the State Oceanic Administration of China (the original strain came from China The Marine Microorganism Culture Collection and Management Center, namely the Third Research Institute of the State Oceanic Administration of China, Xiamen, Fujian, China, www.mccc.org.cn), constructed the genome library of the strain according to the following steps and methods: extracting the deep-sea marine bacterium Janibacter limosus That is, the genomic DNA of the 1C00094 strain of Mirabilis spp. (see below for the specific method), was partially enzymatically digested (the restriction site...
Embodiment 2
[0059] Expression, purification and analysis of embodiment 2 EPSPS protein
[0060] (1) Expression of the target protein EPSPS
[0061] The recombinant plasmid pGEX-6p-1-AroA J.limo Transformed into expression host cell Escherichia coli BL21(DE3). Verify the positive clones, activate the positive clones identified correctly overnight, transfer to 1L LB liquid medium containing 100μg / mL Ampicillin with an inoculation volume of 1% by volume, and culture at 37°C for 2-3h until OD 600 When it reaches about 0.6, the inducer isopropyl-β-D-thiogalactopyranoside (IPTG) is added to a final concentration of 0.5mM, and cultured at 22°C and 180rpm for 25h. The bacteria were collected by centrifugation, and then the pH was adjusted to 7.0 with Hepes buffer (recipe: 50 mM 4-hydroxyethylpiperazineethanesulfonic acid (Hepes), 2 mM dithiothreitol (DTT), and double-distilled water was added to 1 L. Standby) Wash the cells once, suspend them with 50 mL of Hepes buffer, and break the cells wit...
Embodiment 3
[0090] Embodiment 3: Genetic transformation experiment (functional complementation verification)
[0091] The recombinant plasmid pGEX-6p-1-AroA J.limo Transformed into AroA gene-deficient Escherichia coli competent cells (for the transformation process, refer to the transformation process in the above-mentioned Example 1. This bacterial strain does not contain the AroA gene, which can eliminate the interference of the AroA gene contained in general E. coli) (Vaithanomsat and Brown 2007) , were transferred to M9 liquid medium containing 0mM, 50mM, and 100mM glyphosate, respectively, and the growth curve was measured.
[0092] (1) Set comparison
[0093] Primer design and synthesis. According to the Escherichia coli E.coli K-12 AroA gene published by GENEBANK (named AroA E.coli ) nucleotide sequence (accession number: NP_415428.1), the following primer pairs were designed and synthesized:
[0094] Forward primer (5'-AroA E.coli -BamHI):
[0095] 5'-CGC GGATCC ATGGAATCCC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com