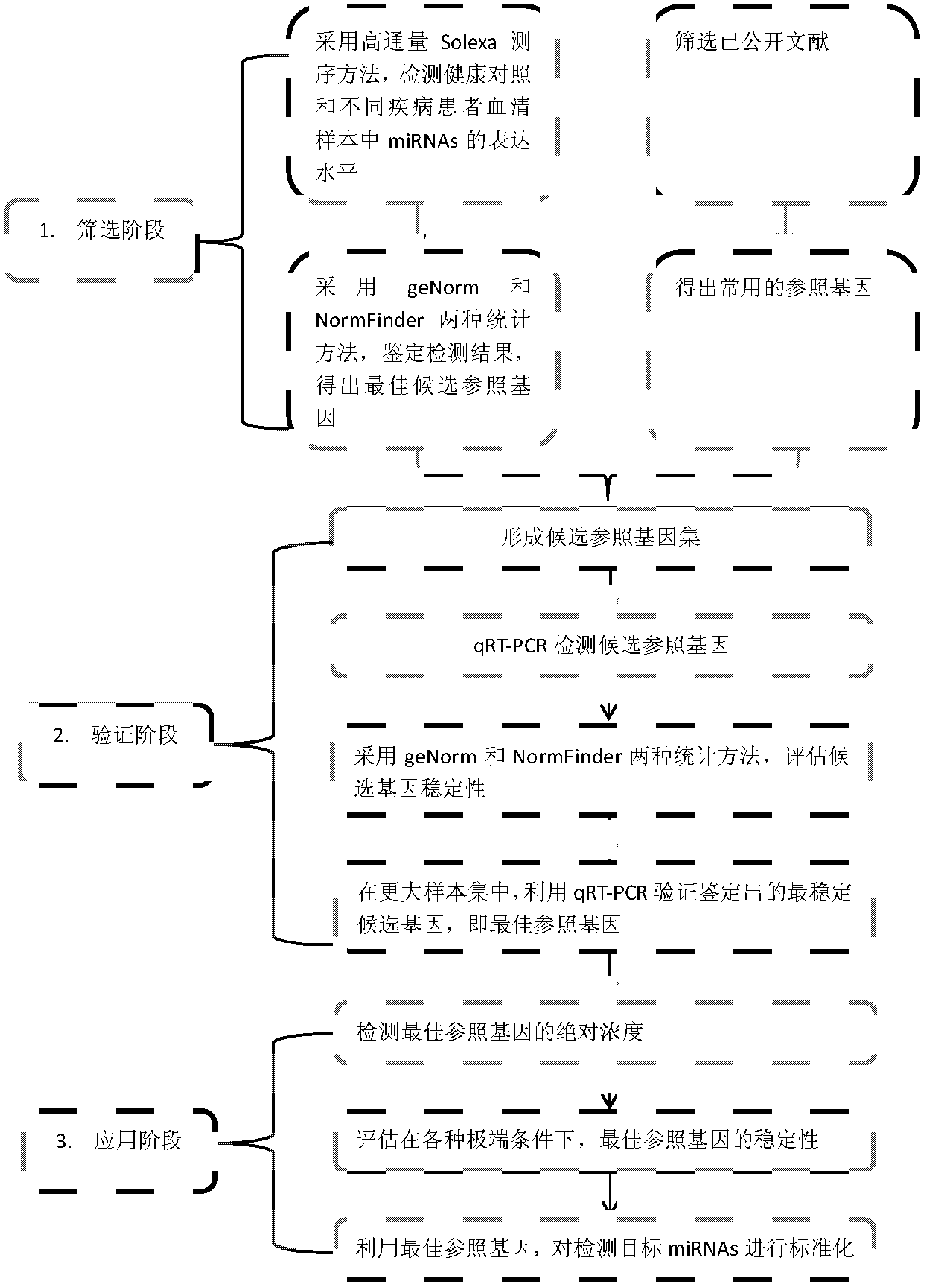
MicroRNA standardization reference gene and application thereof
An internal reference gene and application technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, chemical library, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0088] The preparation of the miRNA chip can adopt conventional manufacturing methods of biochips known in the art. For example, if the solid phase carrier is a modified glass slide or silicon wafer, and the 5' end of the probe contains amino-modified poly dT strings, the oligonucleotide probe can be formulated into a solution, and then spotted with a spotting instrument. The miRNA chip of the present invention can be obtained by arranging in a predetermined sequence or array on a modified glass slide or a silicon chip, and placing it overnight for fixation.
[0089] Reagent test kit
[0090] The present invention also provides a kit comprising a reagent or a chip for determining an internal reference gene or an internal reference gene set, the internal reference gene or an internal reference gene set comprising 1, 2 or 3 microRNAs selected from the following group: let-7d, let-7g, and let-7i;
[0091] Wherein, described reagent is selected from following group:
[0092] (a...
Embodiment 1
[0115] Solexa sequencing and statistical analysis to screen the most stable reference gene
[0116] This example first generates and screens Solexa sequencing data sets to determine stable circulating miRNAs under various physiological and pathological conditions.
[0117] A total of 23 samples were analyzed and miRNAs meeting the following conditions were considered stable:
[0118] (1) expressed in all samples;
[0119] (2) High expression relative to the measured mean;
[0120] (3) The expression level was determined to be stable by measuring the standard deviation.
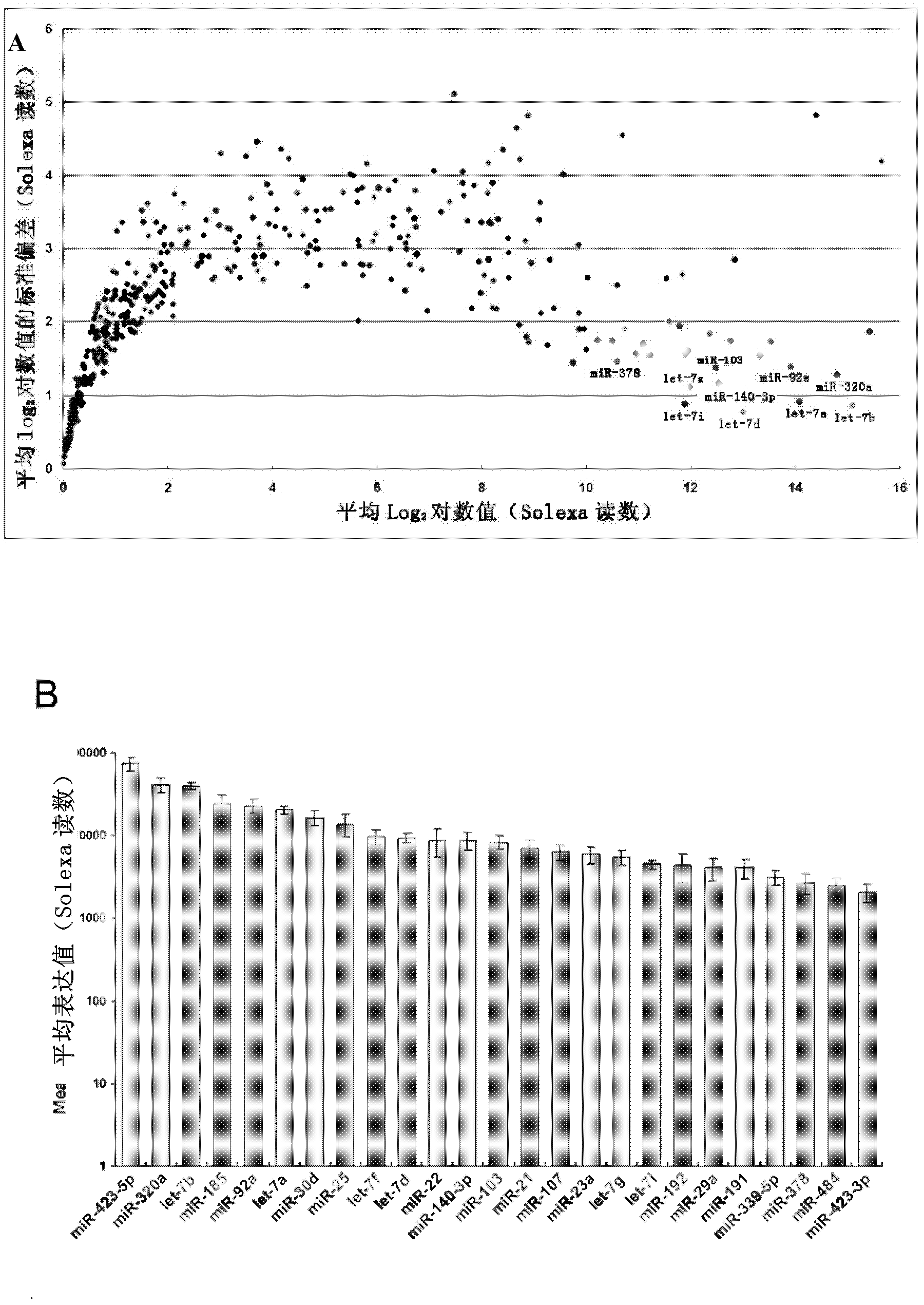
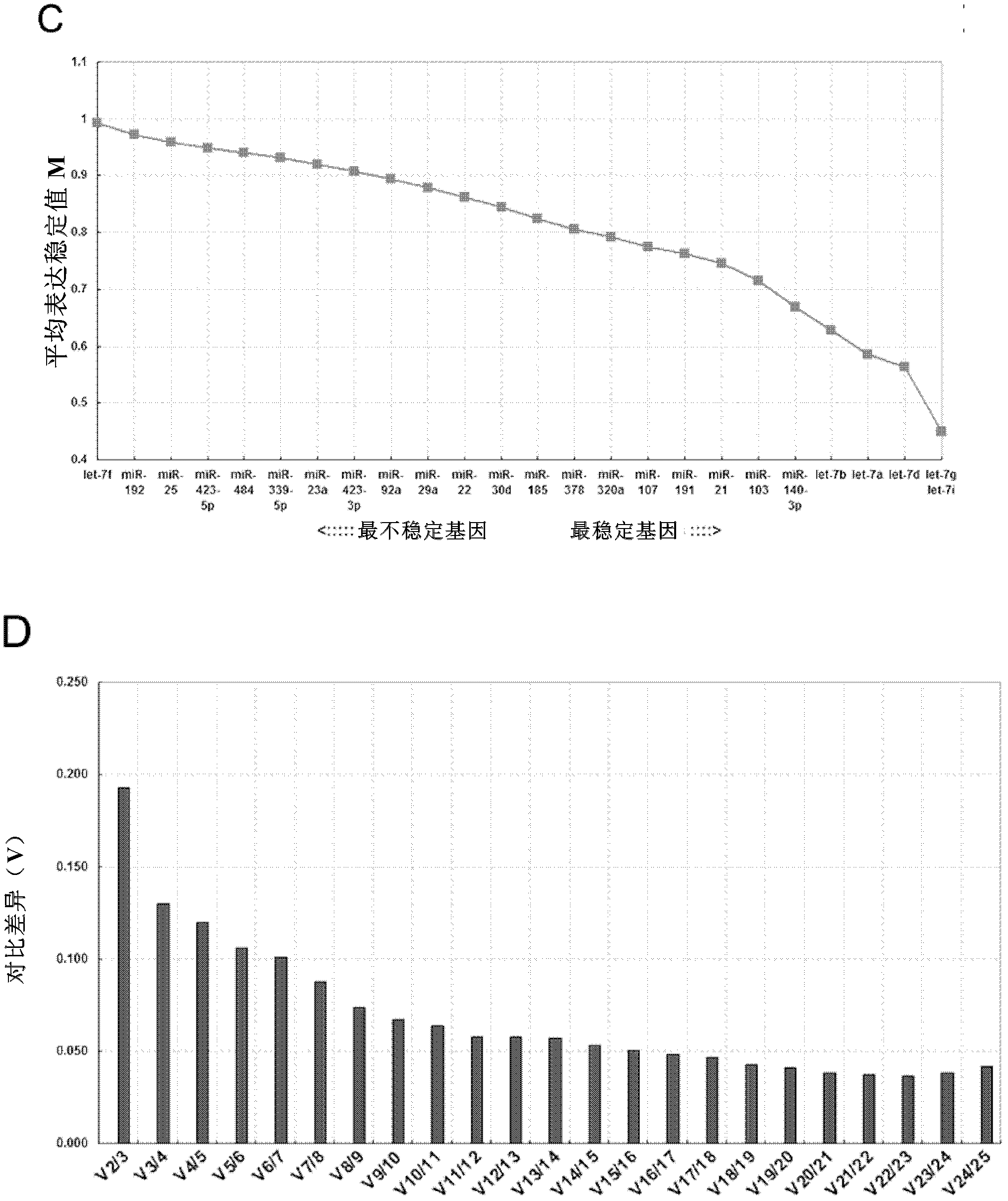
[0121] According to these criteria, 25 miRNAs were screened out as candidate reference genes, the results are shown in figure 2 .
[0122] Such as figure 2 As shown in A, the Solexa reads were converted to logarithmic values, and the genes were ranked by mean expression level and standard deviation; among all the miRNAs screened, 25 miRNAs had higher abundance in the data set (log 2 - Conversion reads ...
Embodiment 2
[0128] Screen for other candidate reference genes
[0129] This example further evaluates and analyzes the three candidate reference genes selected in the previous example and some commonly used reference genes. Commonly used reference genes include macromolecular RNA (GAPDH and β-actin), small nuclear RNA / nucleolus Small RNAs (snRNA / snoRNA) (U6, RNU44, and RNU48) and housekeeping miRNAs (miR-16, miR-191, miR-103, and miR-23a). Based on previous reports on their stable expression in tissues or cells, the inventors selected GAPDH and β-actin. U6, RNU44, RNU48, miR-16, miR-191, miR-103 and miR-23a were selected because they are often used as reference genes when measuring tissue / cell miRNAs. In addition, special attention was given to U6 and miR-16, as they have been used as reference genes for normalization of circulating miRNAs.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com