PRNA mutlivalent junction domain for use in stable multivalent RNA nanoparticles
一种纳米颗粒、稳定的技术,应用在DNA / RNA片段、重组DNA技术、微生物的测定/检验等方向,能够解决RNA纳米颗粒不稳定、阻碍RNA纳米颗粒递送效率和治疗应用、离解等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0214] experimental method
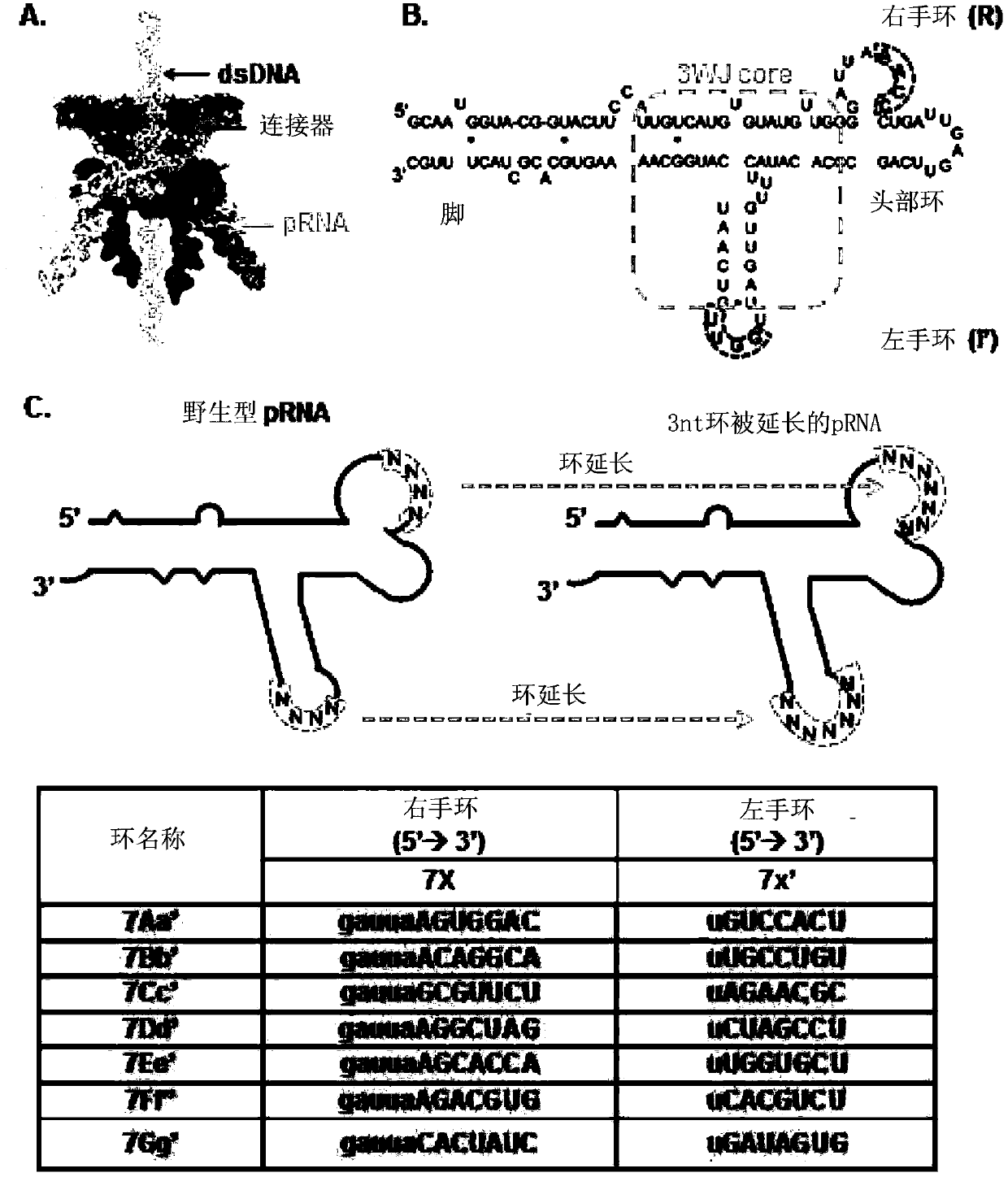
[0215] In vitro synthesis and purification of pRNA and chemically modified pRNA. pRNA was synthesized by enzymatic methods as described previously. RNA oligonucleotides were chemically synthesized by IDT. 2'-Deoxy-2'-fluoro(2'-F) modified RNA was synthesized by in vitro transcription with mutant Y639F T7 RNA polymerase using 2'-F modified dCTP and dUTP (Trilink).
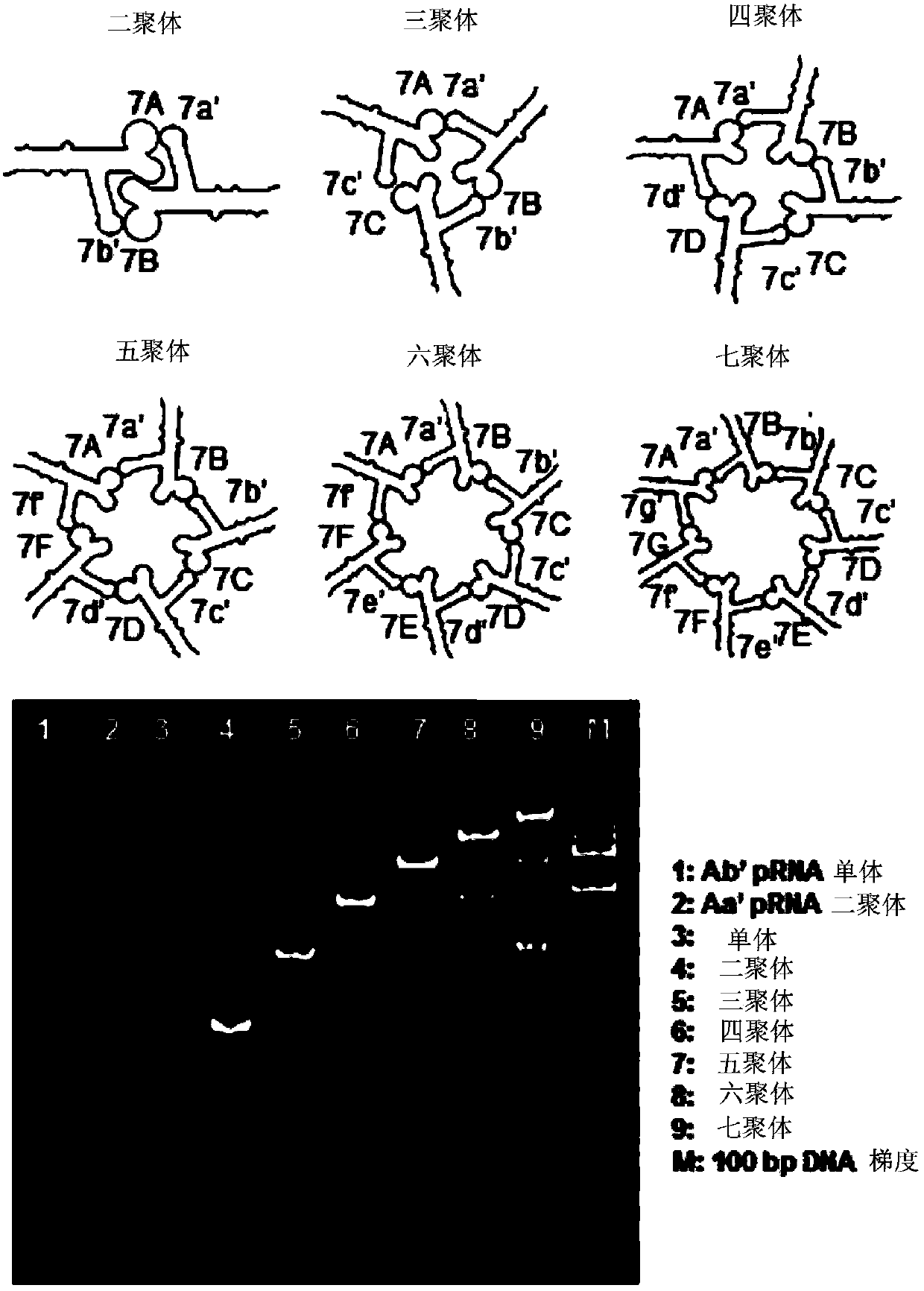
[0216] Construction and purification of pRNA dimers and trimers. In 5mM magnesium [TBM (89mM Tris, 200mM boric acid, 5mM MgCl 2 , pH7.6) or TMS (89mM Tris, 5mM MgCl 2 , pH 7.6) buffer] in the presence of pRNAAb' and pRNABa' in equimolar ratios were mixed to construct dimers. Similarly, trimers were assembled by mixing pRNAAb', pRNABc' and pRNACa'. To purify the complex, the band corresponding to the trifurcation domain was excised from an 8% non-denaturing PAGE gel and washed at 37°C (0.5M NH 4 OAC, 0.1 mM EDTA, 0.1% SDS and 0.5 mM MgCl 2 ) to 4 hours, followed by ethanol precipi...
example 2
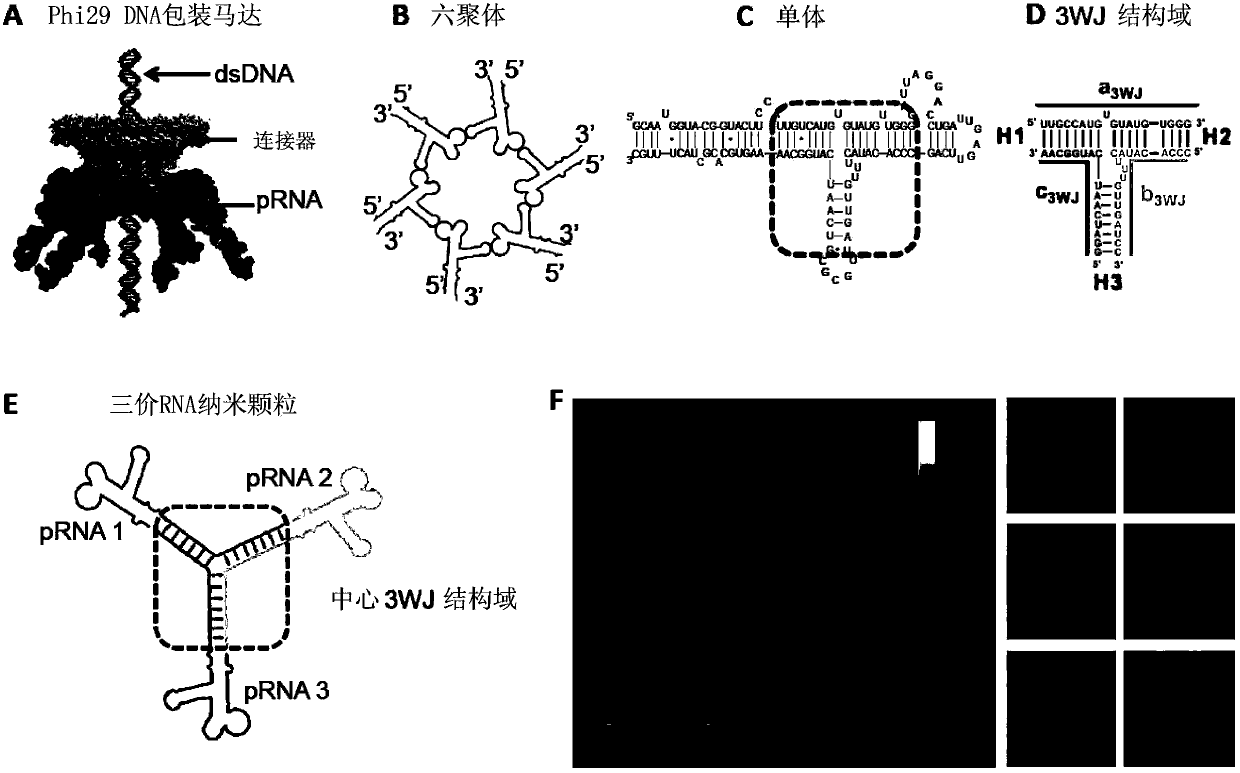
[0265] Construction of trifurcation domain and its derivatives from Phi29pRNA
[0266] Unique properties displayed by trifurcation domains assembled from three RNA oligonucleotides. use a 3WJ , b 3WJ and c 3WJ ( figure 1 D) Three pieces of RNA oligonucleotides representing the construction of the trifurcation domain of phi29pRNA. oligonucleotide a 3WJ , b 3WJ and c 3WJ Two anti-ethidium bromide stains in ( Figure 8 A). Using SYBR Green II RNA gel stain, a 3WJ Can be slightly stained, while c 3WJ is still uncolorable ( Figure 8 A). Ethidium bromide is an intercalating reagent that generally stains dsRNA and dsDNA as well as ssRNA including secondary structure or base stacking. SYBR Green II is known for its ability to stain single- and double-stranded RNA or DNA. especially in c 3WJ In , the absence of staining indicates unique structural properties.
[0267] Three oligonucleotides a 3WJ , b 3WJ and c 3WJ Mixing at room temperature at a stoichiometric rati...
example 3
[0287] Construction of diverse therapeutic RNA nanoparticles using X-motifs as scaffolds
[0288] One advantage of RNA nanotechnology is the feasibility of constructing therapeutic particles with multiple therapeutic agents of defined structure and stoichiometry. However, the controlled assembly of stable RNA nanoparticles with multiple functional molecules to maintain the original role is challenging due to refolding after fusion. In the present invention, we report the construction of thermodynamically stable X-shaped RNA nanoparticles to carry four siRNAs using a redesigned RNA fragment from the central domain of the pRNA of the phage phi29 DNA packaging motor. Tetravalent X-shaped nanoparticles self-assemble very efficiently in the absence of divalent salts, are resistant to denaturation by 8 moles of urea, and remain intact at ultra-low concentrations. We demonstrate that each arm of the four helical strands in the X-motif can contain an siRNA, ribozyme, or aptamer witho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com