Gene chip capable of simultaneously detecting various vibrios and method for detecting vibrios
A gene chip and gene technology, applied in the direction of microbe-based methods, biochemical equipment and methods, microbiological measurement/testing, etc., to achieve efficient and fast detection, high specificity, and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] 1. Probe Design
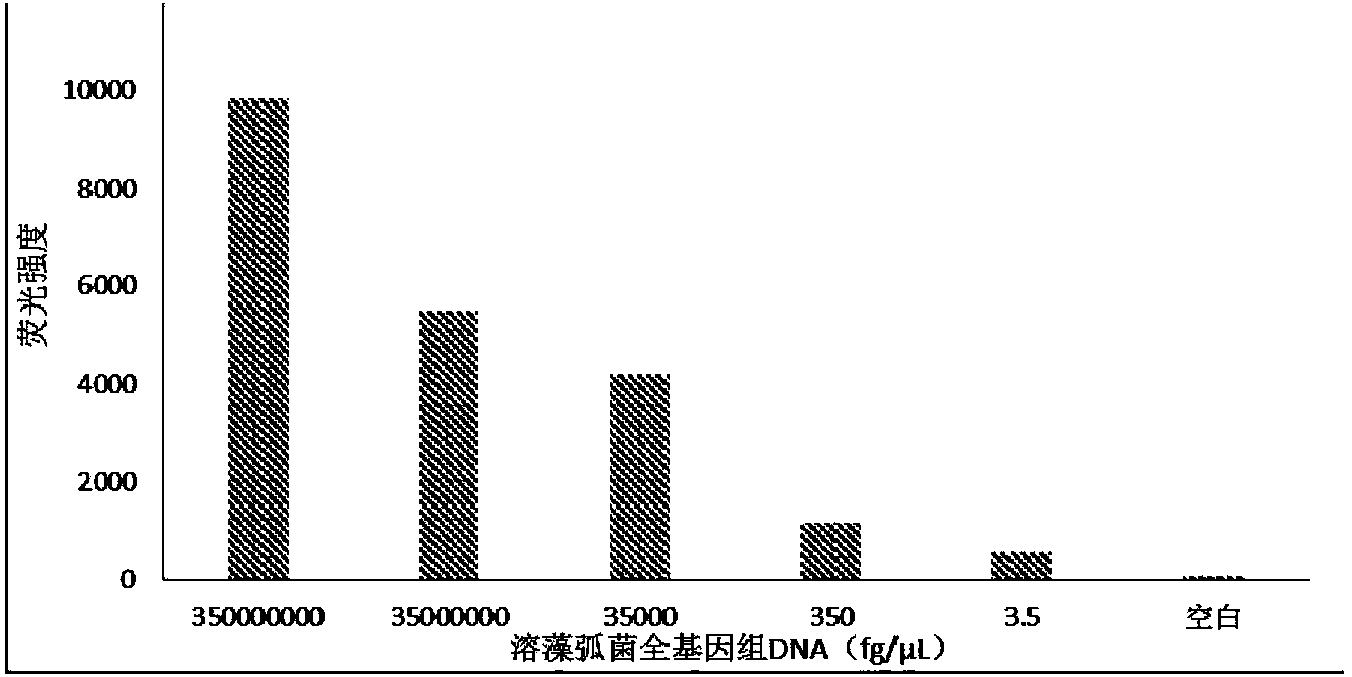
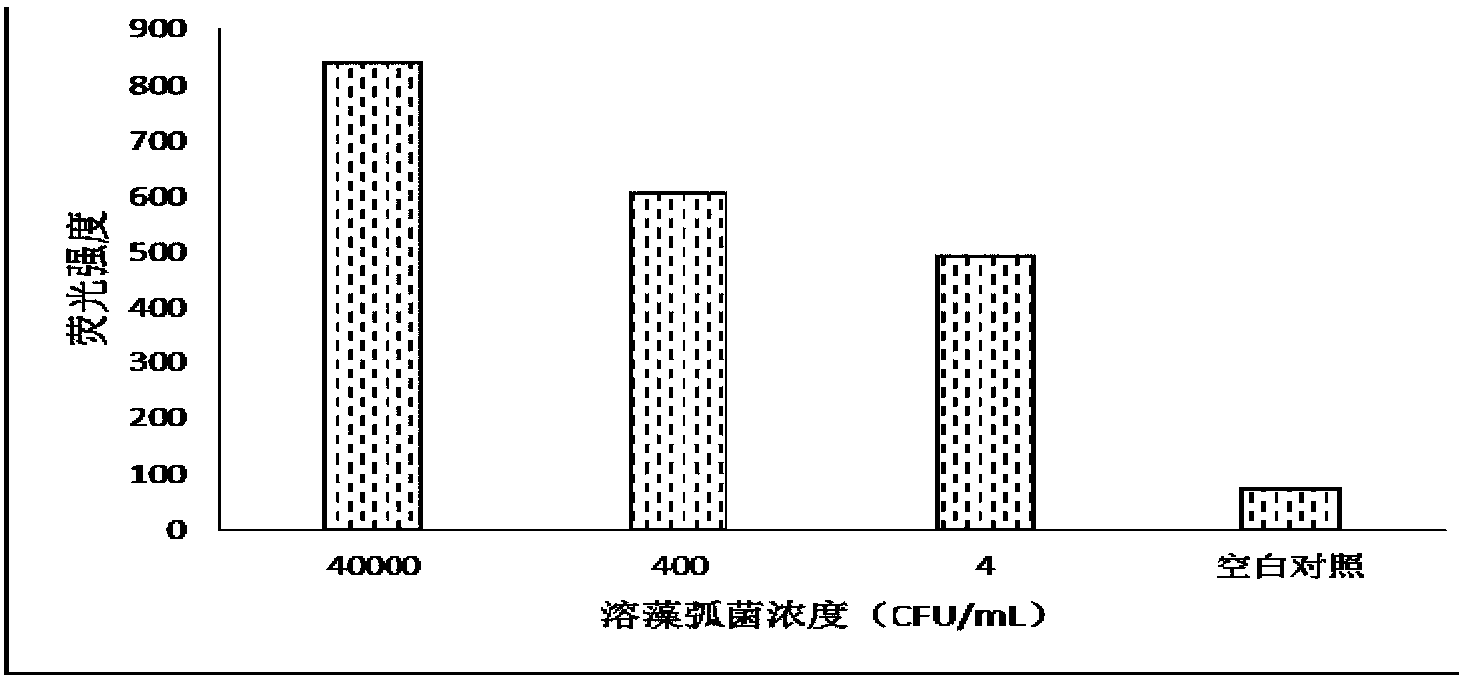
[0042] The gene chip of the invention is used to detect vibrio alginolyticus, vibrio parahaemolyticus, vibrio harveyi and vibrio vulnificus in fishery breeding environment and aquatic products. The probes used are all designed according to the hypervariable region sequences of the hsp60 and toxR genes of the strain, and their specificity is guaranteed by BLAST comparison. table 3. The primers and probes used were commercially synthesized by specialized companies.
[0043] Table 3. Sequence comparison information of the probes applied for in the present invention and similar probes of other published patents
[0044]
[0045] Note: Approximate probe patent publication numbers: 1. CN101475986A; 2. CN101475986A; 3. CN101967510A; 4. CN101967510A; 5. CN101113476A.
[0046] 2. The construction of the gene chip:
[0047] (1) Dilute the commercially synthesized oligonucleotide probe to an appropriate concentration: use 1×TE buffer to dilute to a concent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com