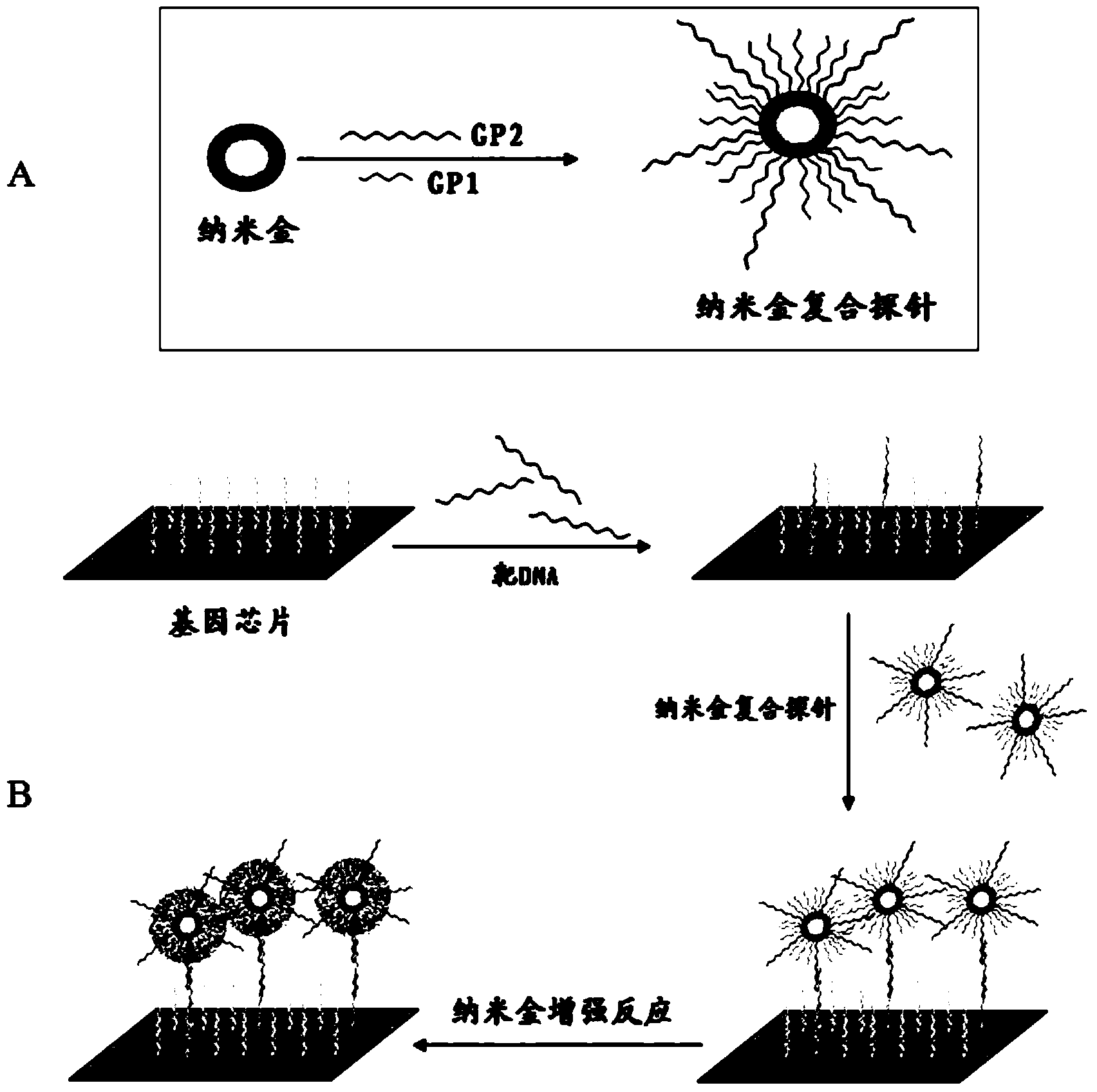
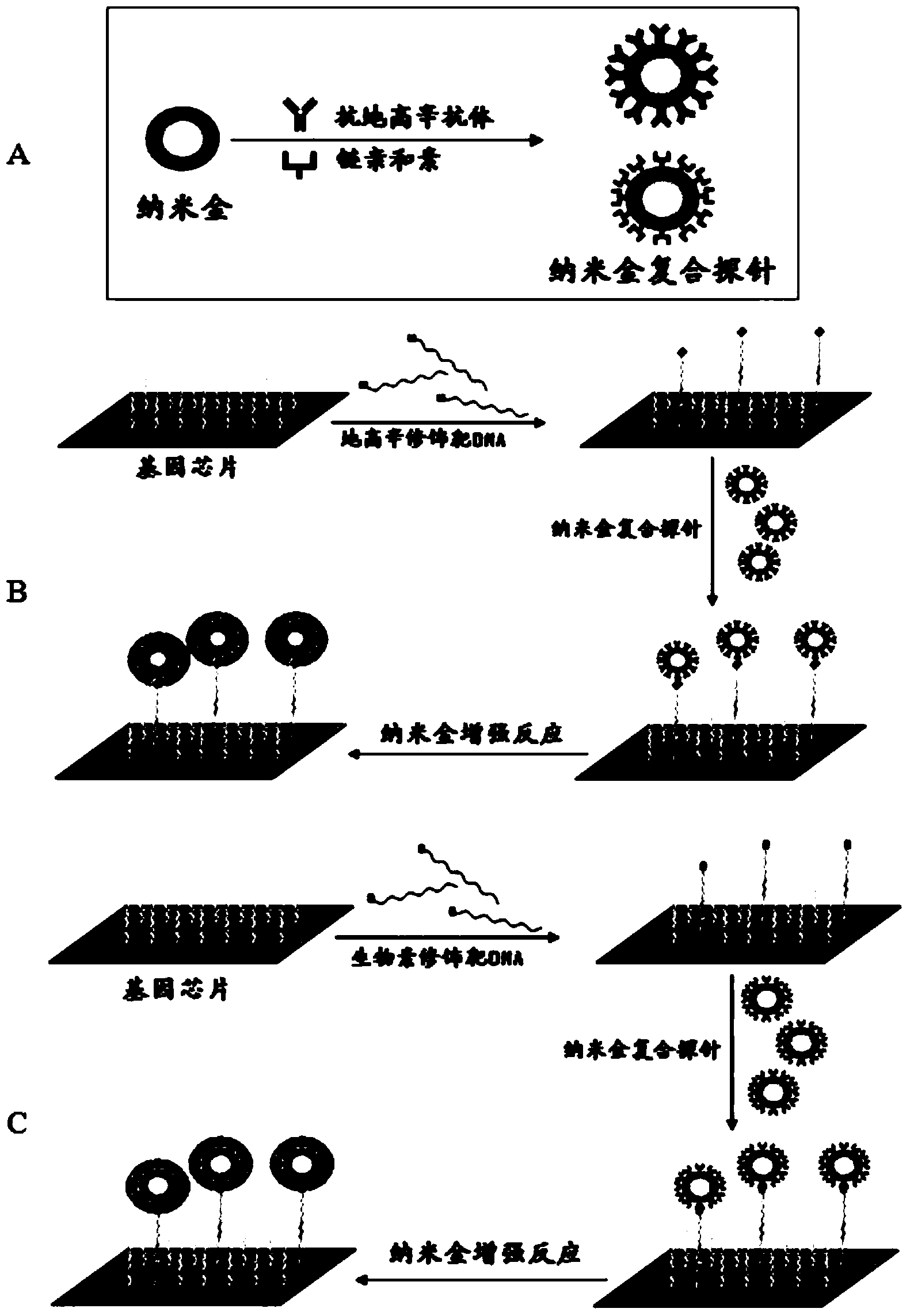
Gold deposition detection method for gene chip
A detection method and gene chip technology, which are applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of limited kit development, high toxicity of silver staining reagents, strong light sensitivity, etc., and improve detection sensitivity. , Simple operation, high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Comparison of the results of detection of Mycobacterium tuberculosis based on hydrogen peroxide reduction and gene chip gold deposition detection methods based on hydroxylamine reduction and silver staining
[0036] 1. Chip Preparation
[0037] Amino-modified probes (see Table 1 for the probe sequence) were dot-patterned on the surface of the aldehyde-modified chip using a chip spotter, and the dot-patterned chip was placed in a closed space with a relative humidity of 75% and a temperature of 30°C for 24 hours. ~48 hours.
[0038] Table 1 Nucleic acid probe sequence list
[0039]
[0040] Note: Biotin: biotin; NH2: amino, all sequences were synthesized by Takara
[0041] 2. Preparation of nano-gold composite probes
[0042] With 0.1mol / L K 2 CO 3 Adjust the pH value of the nano-gold solution (20 μl / ml) to pH 6, and add 1 / 10 volume of streptavidin solution (60 μg / ml) (the concentration is determined by the nano-gold protein stability experiment). Mix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com