Method for marking protein polymer by using quantum dots
A technology for labeling proteins and quantum dots, applied in the field of quantum dot-labeled protein polymers, can solve the problems of poor stability of QDs-labeled proteins, and achieve the effects of high binding stability, solving instability and high repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
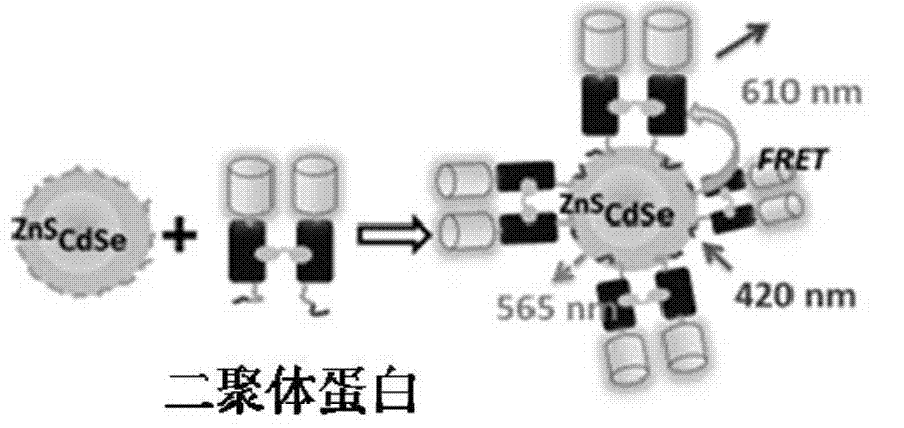
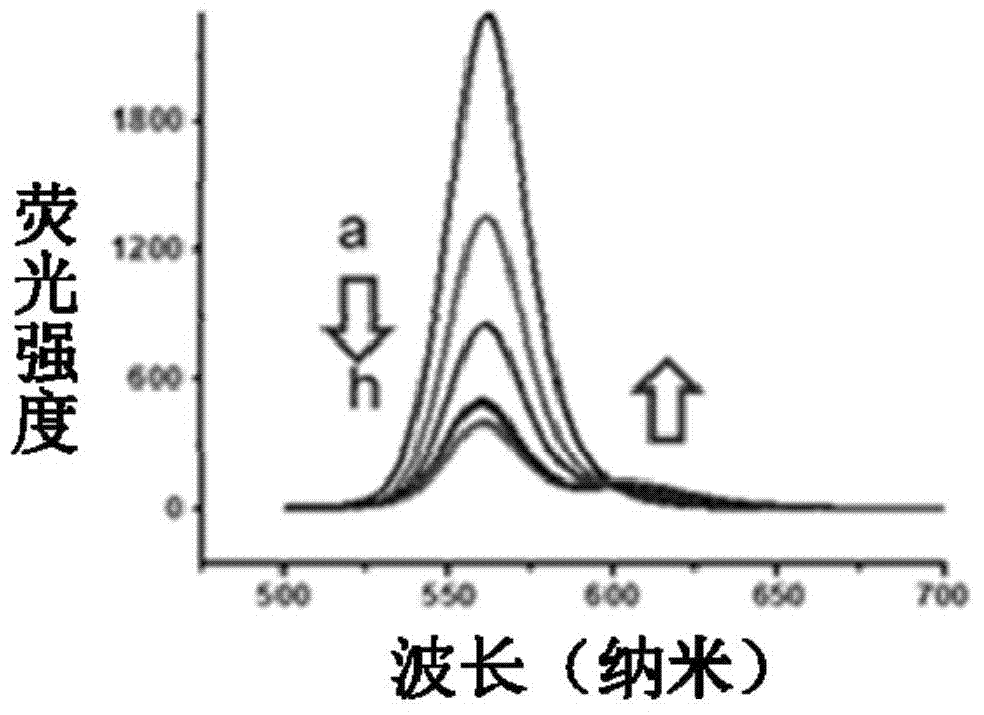
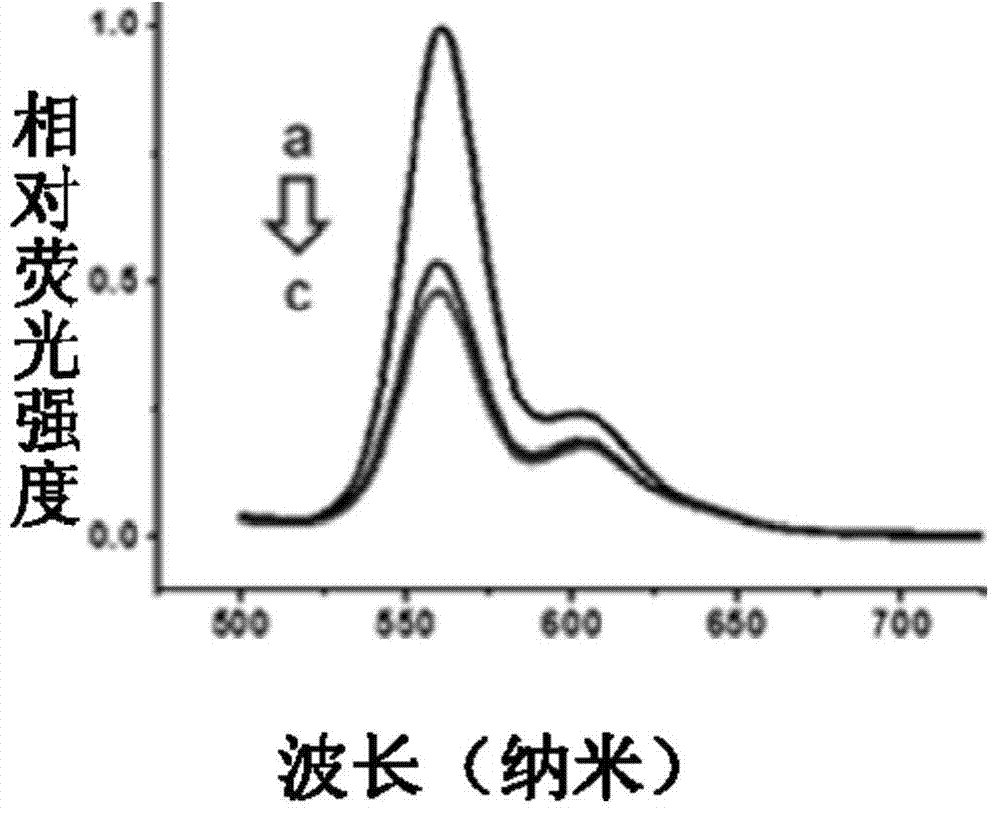
[0020] Select CdSe / ZnS quantum dots with an emission wavelength of 565nm, select TIP 1 Protein, the C-terminus is connected to mCherry, and the N-terminus is connected to a Histag to form a TIP 1 -mCherry monomer. Put two TIP 1 -mCherry monomer via dimeric peptide ligand (CGGWRESAI) 2 connected into a stable H-shape, the TIP 1 -mCherry dimer has 2 Histags ( figure 1 ). Will TIP 1 -mCherry monomer and its dimer are coupled and mixed with quantum dots in different proportions through Histag to obtain TIP 1 -mCherry-QD complex and TIP 1 -mCherry dimer-QD complex for FRET assay ( figure 2 ).
[0021] To detect TIP 1 - Stability of mCherry monomer and its dimer binding to QDs, combining QDs with TIP 1 -After mixing mCherry monomer and its dimer, add imidazole to replace the ligand (final concentration of imidazole is 200mM or 400mM). Imidazole can chelate Zn ions, so it can compete with Histag, and it is possible to bind TIP bound to QDs 1 -mCherry monomer and its dim...
Embodiment 2
[0023] Select CdTe / ZnS quantum dots whose emission wavelength is 612nm, and other steps are the same as in Example 1.
Embodiment 3
[0025] The fusion protein used was TIP 1 -GFP fluorescent protein, other steps are the same as in Example 1.
[0026] It can be seen from the above three examples that the present invention can realize the rapid combination of quantum dots and dimer proteins, and has high stability and convenient operation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com