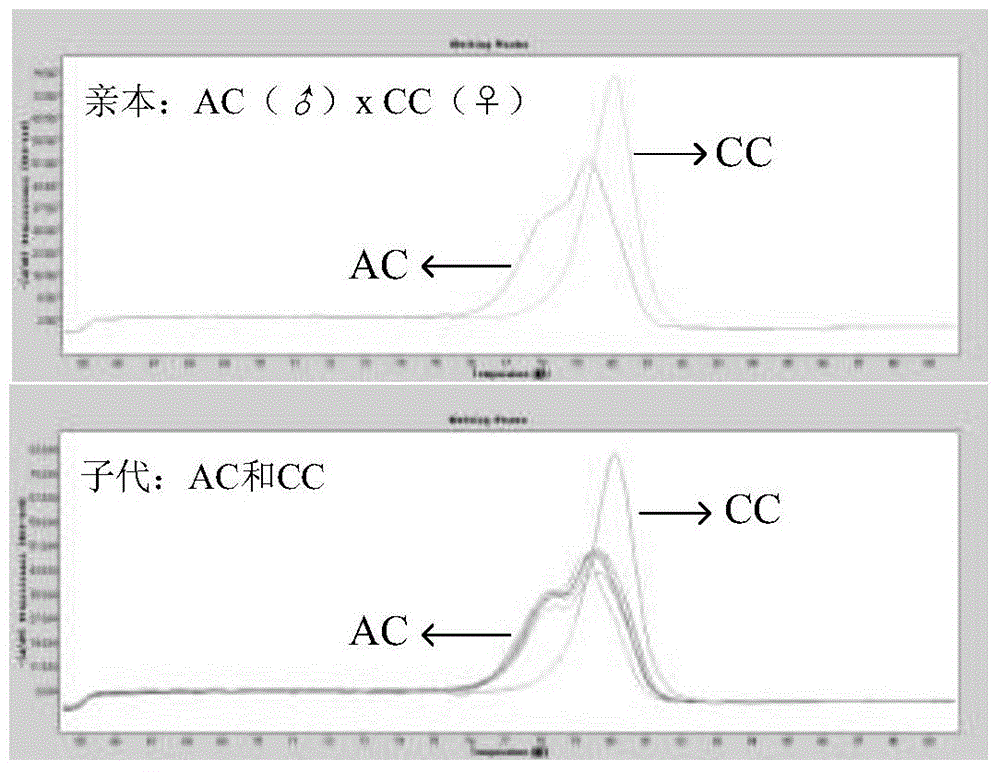
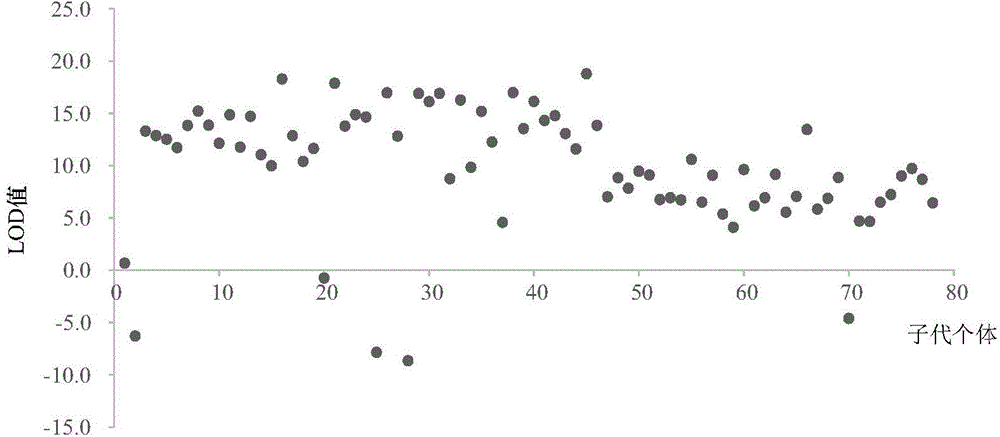
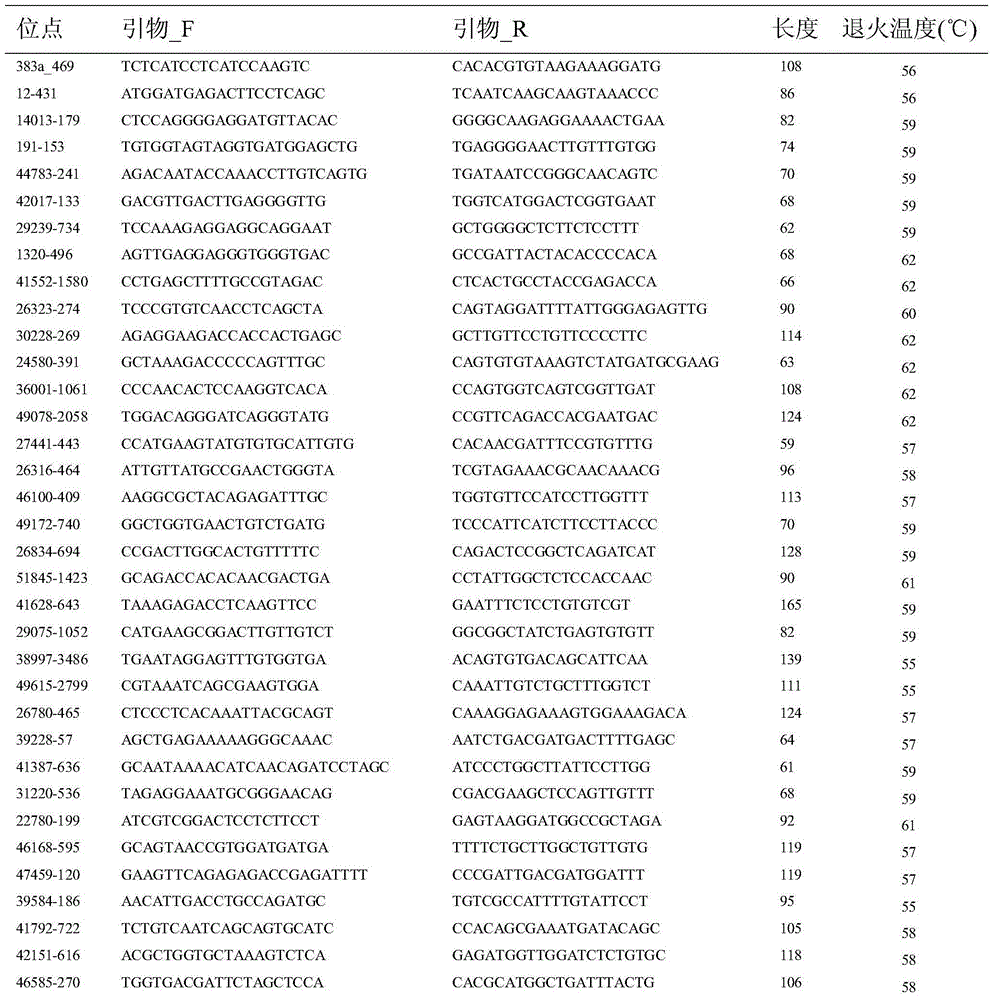
Method applied to identification of pacific oyster family
A Pacific oyster and identification method technology, applied in the field of Pacific oyster family identification, can solve the problems of weak genetic research, influence, lack of improved species, etc., and achieve the effect of high resolution of dissolution curve, high stability and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] 1) Primer design and optimization.
[0022] In the SNP database obtained by the Illumina GoldenGate method, search for the first and second SNP mutation sites (C / T&G / A, C / A&G / T), and use Primer3.0 (http: / / primer3 .ut.ee / ) to design primers. When designing primers, try to avoid hairpin structures and primer dimers. The primer length is 18-30bp, the annealing temperature is 55-65°C, and the length of the amplified fragment is 50-180bp. Genomic DNA of 6 individual Pacific oysters was extracted using Qiagen kits, quantitative analysis of DNA was performed with PicoGreen fluorescent dye kits, and the concentration of sample DNA was adjusted to 5 ng / μL for screening and optimization of SNP markers. Using the quantified DNA as a template, use Roche LightCycler480 for HRM detection, by changing the annealing temperature and Mg 2+ The concentration screened primers with high amplification efficiency and clear melting curve, and obtained the optimal PCR reaction system and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com