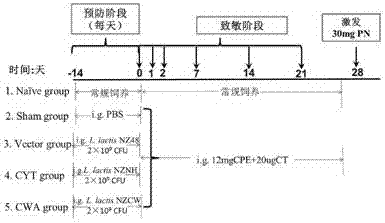
Preparation method and application of recombinant lactococcus lactis capable of displaying peanut allergen on surface
A technology of Lactococcus lactis and peanut allergens, applied in the field of bioengineering, can solve problems such as complex structure, high cost of expression product purification, and limitation of clinical application of Escherichia coli prokaryotic expression system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Codon Optimization of Arah2 Gene Sequence
[0019] First, the signal peptide sequence of Arah2's amino acid sequence (genbank: AAN77576.1) was predicted using the signal peptide prediction software SignalP4.1Server, and the predicted sequence was removed from the gene sequence (genbank: AY158467.1) corresponding to the above amino acid sequence. The signal peptide sequence to obtain the original nucleotide sequence to be optimized.
[0020] The original nucleotide sequence was optimized according to the preferred codon table of Lactococcus lactis, and the optimized gene was named nArah2, which was synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. and subcloned into the vector pUC57 to obtain pUC57 -nArah2 (synthesized by Shanghai Sangon, where pUC57 is a commercial plasmid).
[0021] The natural gene sequence before optimization (genbank: AY158467.1), SEQIDNo.1.
[0022] Optimized gene sequence (nArah2), SEQIDNo.2.
[0023] Amino acid sequence befor...
Embodiment 2
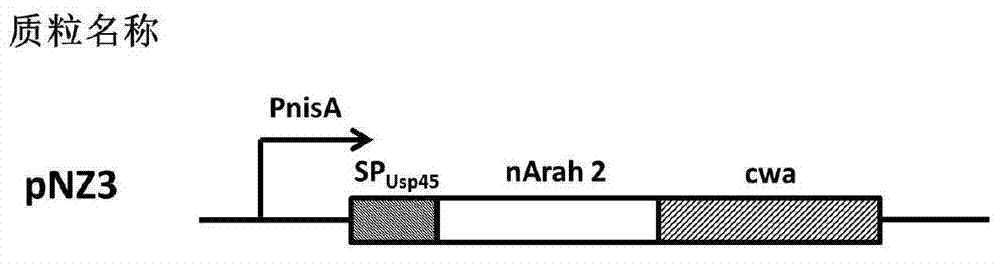
[0025] Example 2 Construction of recombinant expression vector pNZ3
[0026]L.lactissubsp.cremorisMG1363 (Gasson.PlasmidcomplementsofStreptococcus lactis NCDO712and other lactic streptococci after protoplast-induced curing.Journal of Bacteriology.1983,154(1):1-9) inherent N-acetylmuramidase ACMA gene (for Genebank The cA domain in the published gene sequence, GenBank: U17696.1) serves as the anchor function sequence CWA. Using the genomic DNA of L. lactis subsp. cremorisMG1363 as a template, PCR amplification was performed using primers Pca1F: 5'-TAGGGTACCTCTGGTGGCTCGACAACCACAATTAC-3' and Pca1R: 5'-CCGCAAGCTTTTATTTTATTCGTAGATACTGACCAATT-3' PCR amplification-cwa sequence. The PCR reaction system (total volume: 50 μL) is: 10×KODplus buffer: 5 μL; dNTP: 5 μL; MgSO4: 2 μL; DNA template: 2 μL; upstream primer / downstream primer (20 μM): each 1 μL; KODplus: 0.5 μL; ddH 2 O: 33.5 μL. Reaction program: 95°C for 30s (denaturation), 59°C for 30s (annealing), 68°C for 60s (extension), 3...
Embodiment 3
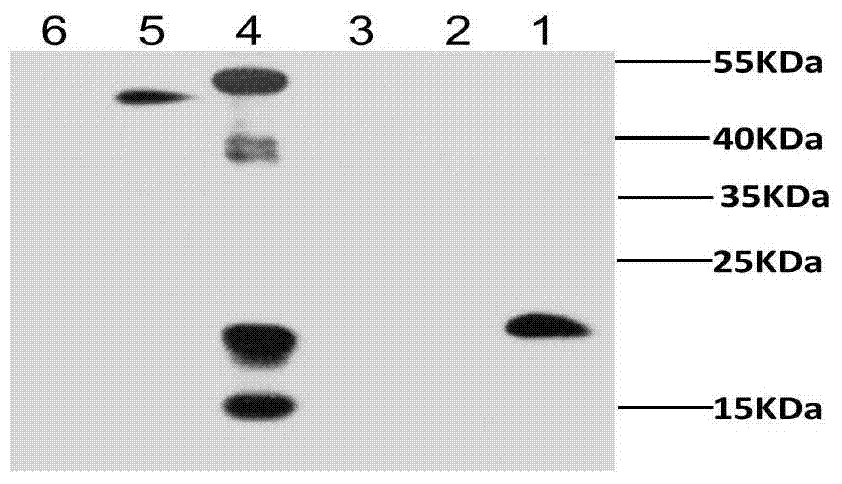
[0027] Example 3 Preparation of recombinant Lactococcus lactis
[0028] The above constructed plasmid pNZ3 was electroporated to transform L.lactisNZ9000 (Kuipers etal. Quorum sensing-controlled gene expression in lactic acid bacteria. Journal of Biotechnology. 1998, 64(1): 15-21) competent cells. Pick a single colony for colony PCR and double enzyme digestion verification, and send it to Sangon Bioengineering (Shanghai) Co., Ltd. for sequencing verification. The sequenced correct transformant is named L.lactisNZCW, and the empty plasmid strain L.lactisNZ9000 / pNZ8148 will be carried Named L. lactisNZ48.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com