Saccharopolyspora spinosa genome scale metabolic network model and construction method and application thereof
A technology of Saccharopolyspora spinosa and genome scale, applied in special data processing applications, instruments, electronic digital data processing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] A method for constructing a genome-scale metabolic network model of Saccharopolyspora spinosa, comprising the steps of:
[0029] According to the annotation information of the genome sequence of Saccharopolyspora spinosa in the KEGG and NCBI databases, the characteristic reactions of spinosyn biosynthesis (see Table 1) and bacterial synthesis reactions (see Table 2) were added:
[0030] Table 1 spinosad biosynthesis reaction
[0031]
[0032]
[0033] Table 2 bacterial synthesis reaction
[0034]
[0035]
[0036] And manually refine the network reaction (removing redundant reactions, adding reaction cofactors, adjusting the reversible direction of the reaction, balancing the reaction, adding transport reactions, adding exchange reactions, analysis to fill in the gaps), manual refining details see (Nature protocols.A protocol for generating a high-quality genome-scale metabolic reconstruction. 2010.) Obtained the genome-scale metabolic network model of Saccha...
Embodiment 2
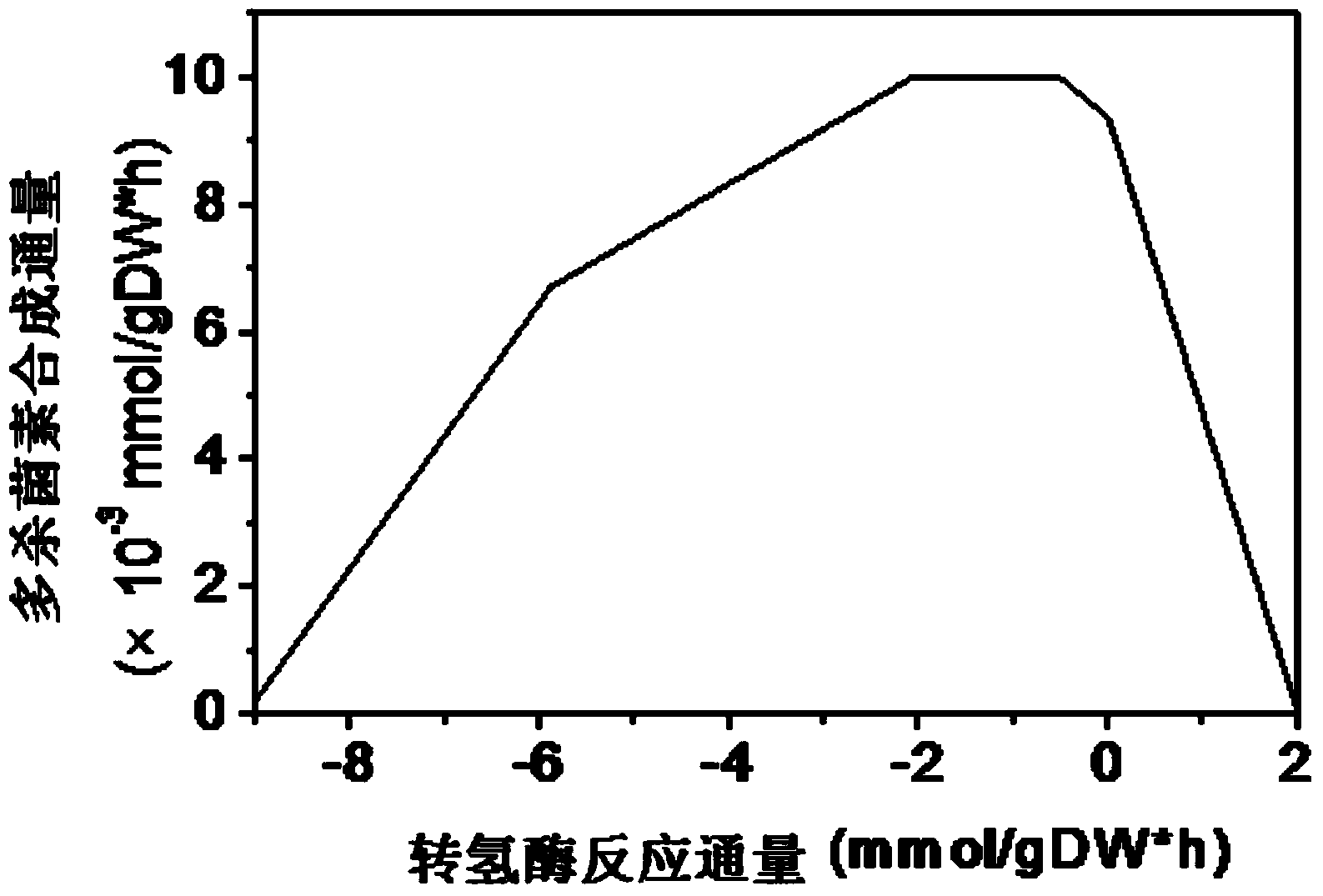
[0043] The Saccharopolyspora spinosa genome-scale metabolic network model constructed in Example 1 is converted into a computer-recognizable mathematical coefficient matrix, and the Matlab software platform is used to catalyze the reaction with the transhydrogenase PntAB by means of the robustness analysis tool in the COBRA toolbox. In order to control the reaction, the synthesis reaction of spinosyn was taken as the target reaction, and the effect of the gene activity of transhydrogenase PntAB on the synthesis rate of spinosyn was determined. The simulation results were as follows: figure 1 .
Embodiment 3
[0045] Molecular amplification is carried out to the transhydrogenase gene pntAB determined in Example 2, and primers are designed according to the transhydrogenase gene sequence on NCBI (the underline represents the restriction site):
[0046] Upstream primer pntAB-F:CCTA TCTAGA CGCCGGAACAAGGACG (shown in SEQ ID NO.1)
[0047] Downstream primer pntAB-R:GGATC CATATG ACCTCTCCGGAGAGC (shown in SEQ ID NO.2)
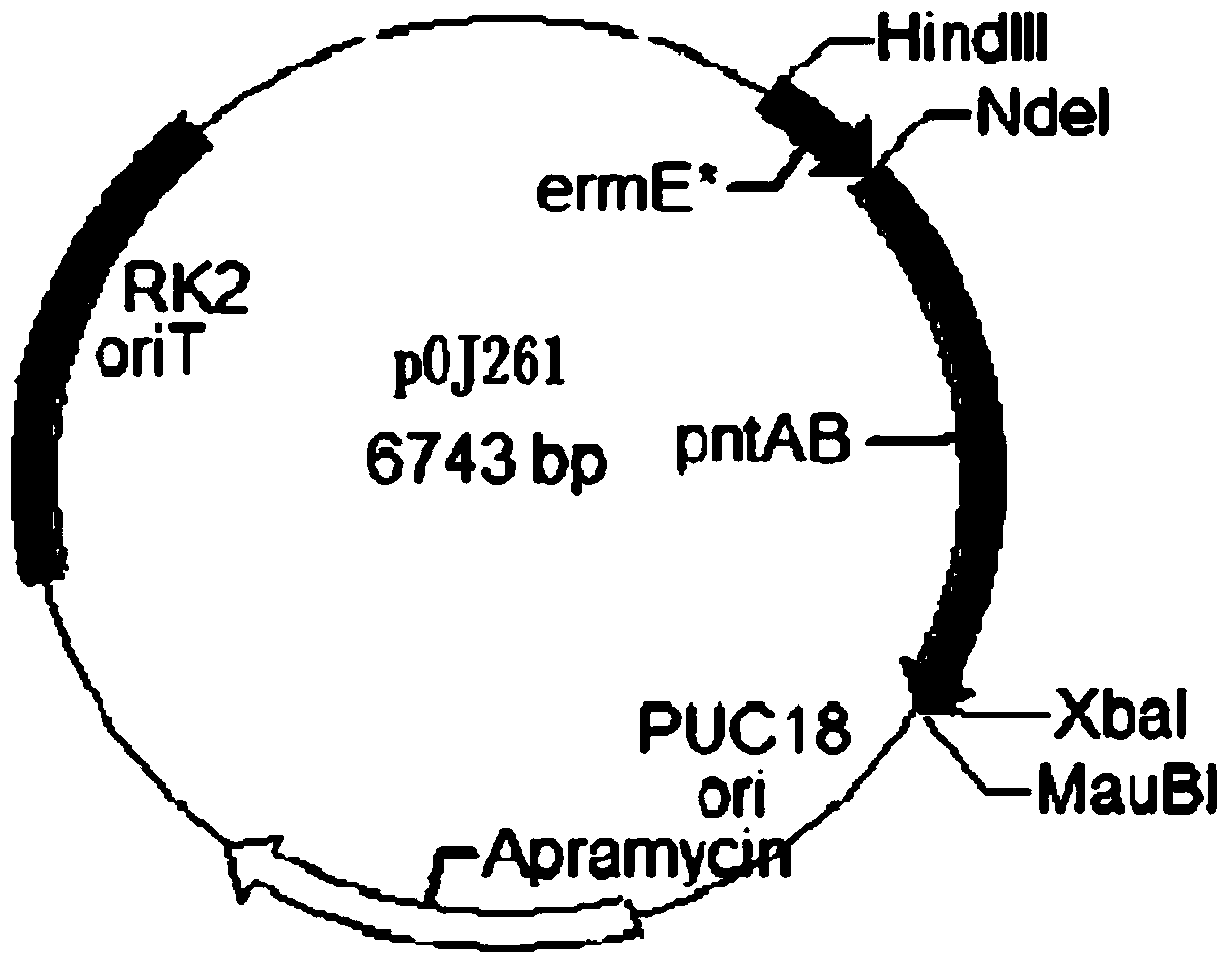
[0048] Using this pair of primers, using the Saccharopolyspora spinosa genome as a template, the transhydrogenase PntAB gene was amplified by PCR technology, and the transhydrogenase pntAB gene was inserted into the Streptomyces strong Promoter ermE* on plasmid pOJ260, resulting in plasmid pOJ261 (see figure 2 ), transform Escherichia coli DH5а, extract the plasmid and transfer it into Escherichia coli ET12567 (ATCC product number: BAA-525), and transfer it into Saccharopolyspora spinosa (Saccharopolyspora spinosa ATCC49460) by the combined transfer method, with 50 μg / m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com