Method for detecting miRNA (micro Ribose Nucleic Acid) by improved stem-lop primer qRT-PCR (Quantitative Reverse Transcription Polymerase Chain Reaction)
A technology of stem-loop primers and detection primers, which is applied in the direction of biochemical equipment and methods, and the determination/inspection of microorganisms. It can solve the problems of high cost, inability to fully guarantee the specificity of cDNA sequences, and the inability to analyze the specificity of PCR reactions. High accuracy and precision, technology cost savings, good sensitivity results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Design of miRNA detection primers
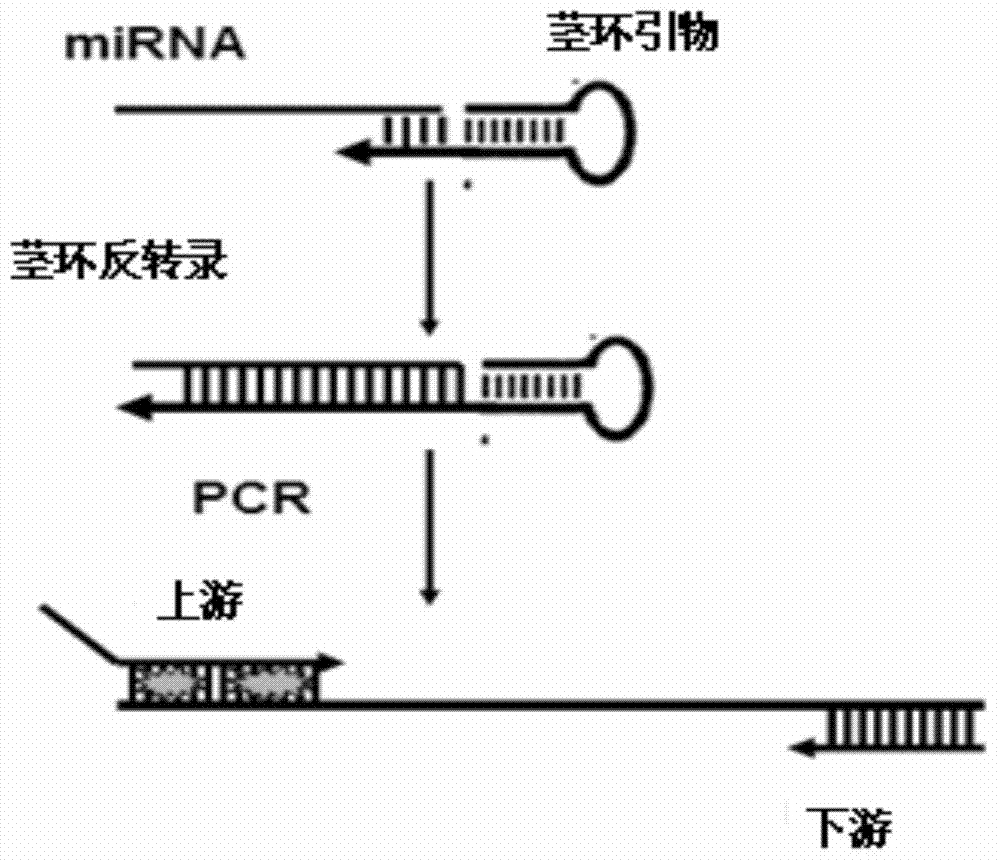
[0042] (1) Stem-loop primers consist of two parts, a universal stem-loop sequence and a specific primer sequence complementary to the 3'end of the target miRNA; the optimized stem-loop primer increases the number of bases that complement the target miRNA from 7bp to 11bp.
[0043] (2) The downstream primers of qPCR are aimed at the universal stem-loop sequence, and the upstream primers are aimed at the 5'end of the target miRNA, and the 5'end has an 8bp tail. The primer design is as follows figure 2 Shown.
[0044] 2. Verify the specificity and sensitivity of primers
[0045] (1) Reverse transcription
[0046] SuperScript II Reverse Transcriptase Kit (Fermentas) was used for reverse transcription. The final reaction system 10μL includes: 6.25μL DNase I-treated total RNA, 0.5μL stem-loop primer (2μM), 0.5μL dNTP Mix (10mM). First, the reaction was carried out at 16°C for 15 minutes, and then the remaining system (2μL 5×RT buffer, 0.5μL Reve...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com