Method for separating and regulating peanut embryonic development gene
A separation method and embryo development technology, applied in biochemical equipment and methods, microbial measurement/inspection, DNA preparation, etc., can solve the problems of unclear molecular mechanism and achieve the effect of simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
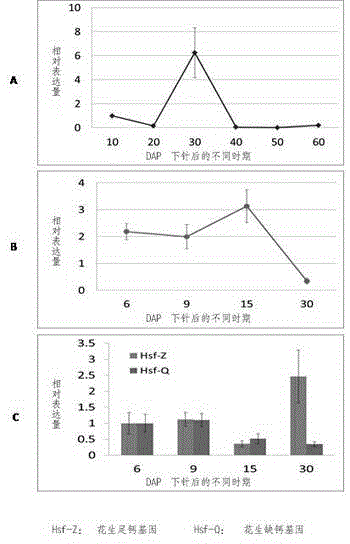
[0022] [Example 1] Extraction of Total RNA in Early Embryos of Peanut Deficiency and Sufficiency Calcium
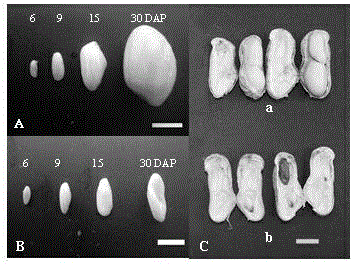
[0023] The fine peanut variety Minhua No. 6 bred by Fujian Agriculture and Forestry University was used as the material, and a typical calcium-deficient peanut empty pod producing area was selected in Pingtan County, Fujian Province. The exchangeable calcium content in the soil was 49.54 ug / kg. For calcium deficiency treatment, on this basis, 75kg of lime is applied per mu, and the exchangeable calcium content in the soil reaches 129.61 mg / kg (the exchangeable calcium content in the soil that causes calcium deficiency in peanuts is 100mg / kg), which is used as sufficient calcium. deal with. Under the conditions of calcium deficiency and sufficient calcium, the young fruits of peanuts on the 6th, 9th, and 15th days after the needles were inserted into the soil were removed, and the immature embryos were quickly thrown into liquid nitrogen and stored at -80°C for later us...
Embodiment 2
[0024] [Example 2] Construction of suppression subtractive hybridization library
[0025] The suppression subtractive hybridization library of peanut early embryos under calcium deficiency and sufficient conditions was constructed according to the PCR-SelectTM cDNA Subtraction Kit (BD Biosciences-Clontech, Palo Alto, CA). When the early peanut embryos under the calcium deficiency condition were the test group, the peanut early embryos under the sufficient calcium condition were the driving group; on the contrary, when the peanut early embryos under the calcium deficiency condition were the detection group, the early peanut embryos under the calcium deficiency condition were the driving group. Take 1 μg of mRNA from the detection group and the driver group respectively and carry out reverse transcription to synthesize cDNA according to the M-MLV RTase cDNA Synthesis Kit (Takara, Japan). Rsa Ⅰ Digest at 37°C for 1.5 h, then ligate linker 1 (5'-CTAATACGACTCACTATAGGGCTCGAGCGGCCGC...
Embodiment 3
[0026] [Example 3] Calcium Deficiency Induced Early Peanut Embryo Suppression Subtractive Hybridization Library Screening
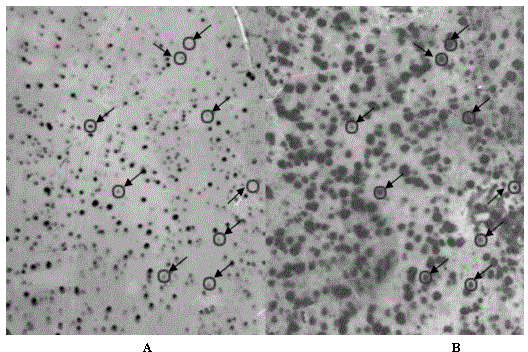
[0027] 2 μg of depleted double-stranded cDNA of peanut early embryos under calcium-deficient and sufficient conditions were taken respectively, and probe-labeled with DIG-dUTP (Roche) according to DIG High Prime DNA Labeling and Detection Starter Kit II (Roche). Take 1 μl of the amplified library stored at -70°C and dilute to 10 with LB liquid medium 3 , 10 4 , 10 5 , take 1 μl respectively, add 100 μl liquid LB, smear on the LB solid medium plate containing 100 μg / ml ampicillin, after culturing overnight, calculate the number of grown clones. Spread the Hybond-N+ nylon membrane (Amersham) on the LB plate containing 100 μg / ml ampicillin, and try to avoid generating air bubbles. According to the titer detection results, take an appropriate amount of library bacteria solution according to 2000 clones per plate and evenly smear the plate, dry it on the ul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com