SNP (single-nucleotide polymorphism) marker related to Crassostrea gigas glycogen content character and application thereof
A long oyster and glycogen technology, which is applied in the fields of molecular biology and genetics and breeding, can solve the problems such as rare research on the quality traits of the long oyster, and achieves the effects of avoiding the use of concentrated acid and alkali, and being safe and easy to operate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
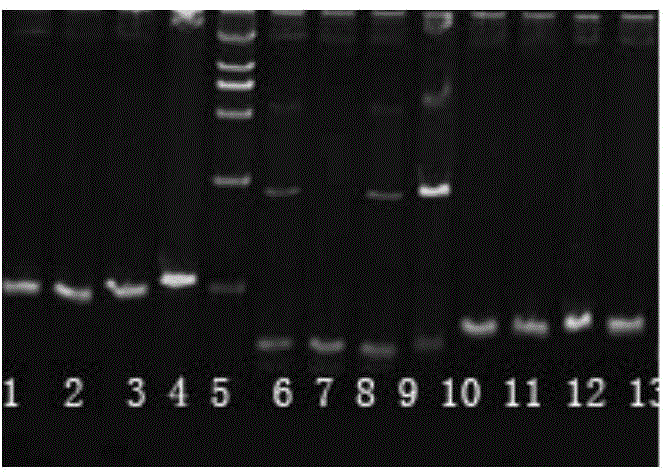
[0033] Screening of SNP sites related to glycogen content
[0034] a) Collection of samples: A total of 144 wild oysters were collected from wild oyster populations in Jiaonan, Qingdao. They were dissected, and samples were taken by tissue (adductor muscle, gill tissue, etc.), and frozen in liquid nitrogen at -80°C. Save for later.
[0035] b) Detection of glycogen content: the relative content of glycogen in the adductor muscle of each individual was detected with the muscle glycogen and liver glycogen determination kit of Nanjing Jiancheng Bioengineering Institute, and the detection method was carried out according to the operation steps in the manual.
[0036] 1) Sampling: Take the frozen muscle sample, rinse it with normal saline, blot it dry with filter paper, and weigh it (sample weight ≤ 100 mg).
[0037] 2) Hydrolysis: according to the weight of the sample (mg): the volume of lye in the kit (μl) = 1:3, add them together to the test tube, cook in a boiling water bath f...
Embodiment 2
[0070] Application of a SNP marker associated with glycogen content of long oyster
[0071] a) Collection of samples: A total of 96 wild oysters were collected from Shentanggou, Qingdao. They were dissected, gills and adductor muscles were taken, and they were quickly frozen with liquid nitrogen and stored at -80°C for later use.
[0072] b) DNA extraction: the extraction method is the same as c) in Example 1.
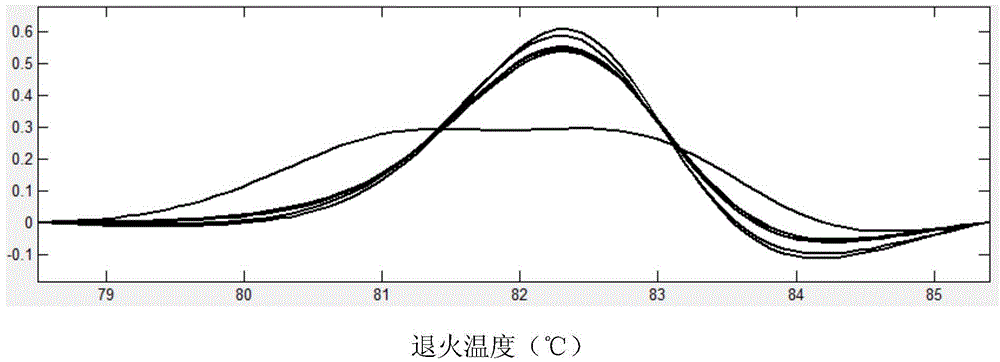
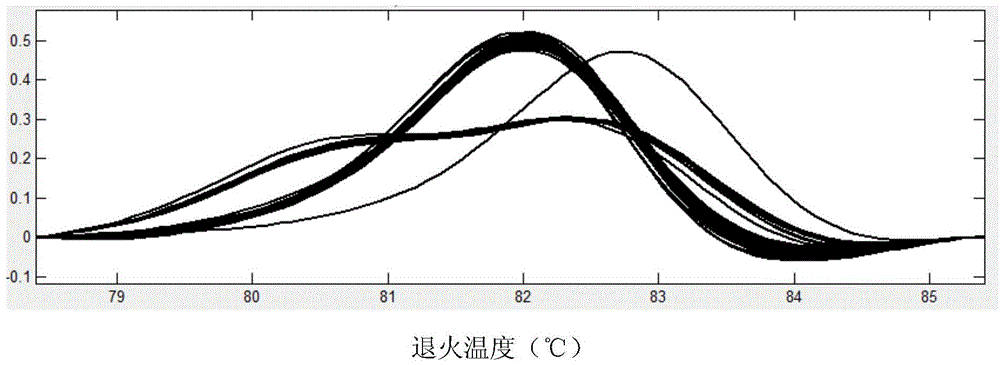
[0073] c) SNP site TY202 genotype detection:
[0074] 1) Using the DNA of 96 wild long oysters in Shentanggou as a template, PCR amplification was carried out with specific primers of TY202, and the amplification reaction was carried out in a skirted 96-well PCR reaction plate. Seal with mineral oil, and the reaction conditions are shown in Table 2.
[0075] 2) After the reaction, add 1 μl of internal standard (internal standard as above) and 1 μl of LC-green dye, denature at 95°C for 10 minutes after transient centrifugation, and cool to room temperature.
[0076] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com