Nucleotide sequence, molecular probe and method for discriminating paphiopedilum micranthum
A technology of nucleotide sequence and P. sclera, applied in biochemical equipment and methods, determination/inspection of microorganisms, DNA/RNA fragments, etc. Simple, low sample volume, short time effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
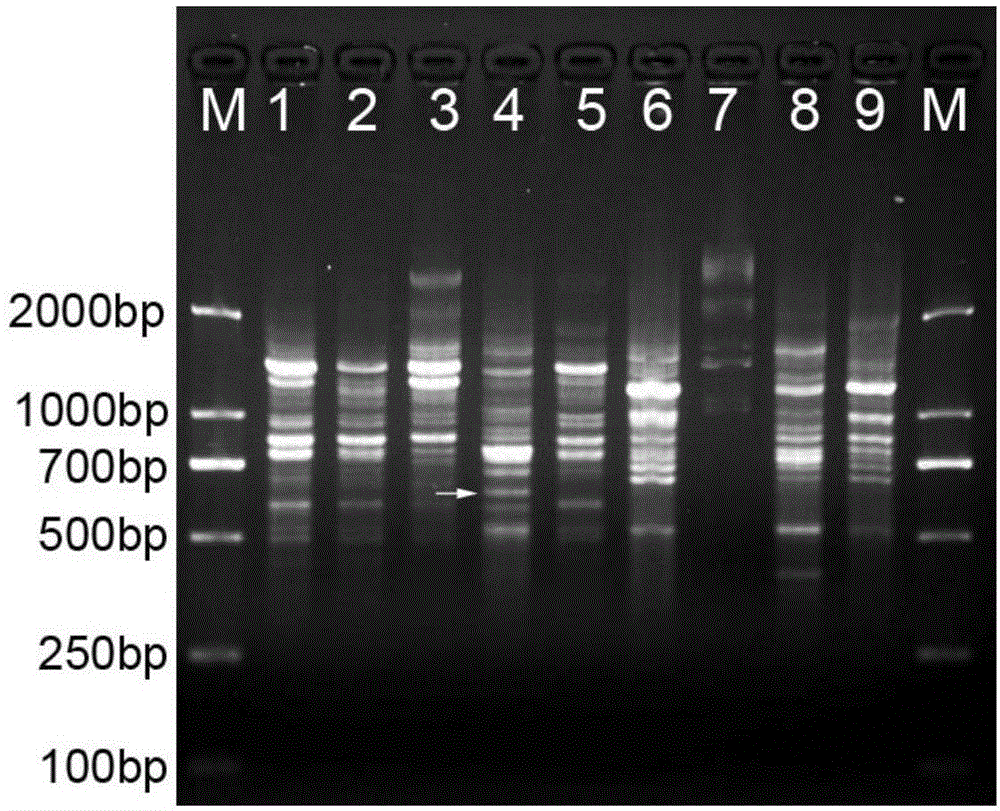
[0026] Example 1: Preparation of the specific nucleotide sequence of Paphiopediphyllum
[0027] 1. Extraction of Genomic DNA
[0028] Cut 0.2 g of the leaves of Paphiopedilum preserved at -80°C, put them into a mortar, immediately add liquid nitrogen to grind to powder, and then use the UNIQ-10 column plant genomic DNA extraction kit of Shanghai Sangon Bioengineering Co., Ltd. to extract Genomic DNA of the genus Paphiopedilum was detected by electrophoresis with 1% agarose gel, and the DNA concentration was detected with a UV spectrophotometer, and diluted to 50ng / μl.
[0029] 2. SCoT–PCR reaction and electrophoresis detection
[0030] Use SCoT universal primer 2 (5'-CAACAATGGCTACCACCC-3') for PCR amplification, the amplification system is: 2μl 10×Buffer, 2μl MgCl 2 (25mM), 0.8μl dNTPs (10mM), 1μl SCoT primer 2 (10μM), 1μl template DNA (50ng / μl), 0.5μl Taq enzyme (2U / μl), 12.7μl ddH 2 O. The total volume is 20 μl.
[0031] The PCR program was: pre-denaturation at 94°C f...
Embodiment 2
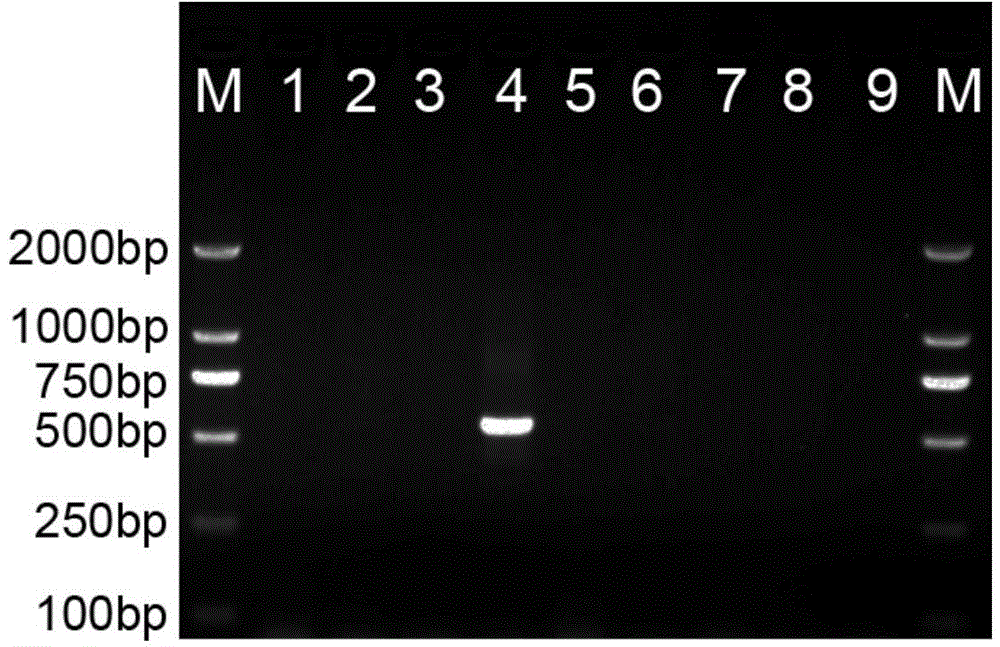
[0035] Example 2: Preparation of P. sclerophyllum-specific nucleotide molecular probe YYF / YYR, PCR reaction and electrophoresis detection
[0036] On the basis of obtaining the specific nucleotide sequence of P. sclerophyllum, use the Primer Primer 5.0 software to design the nucleotide sequence of YYF / YYR (respectively shown in SEQ ID NO.2 and SEQ ID NO.3) , primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd. Then use the designed and synthesized primers YYF / YYR to amplify and detect different Paphiopedilum samples (see the description for details).
[0037] The PCR amplification system is 2μl 10×Buffer, 2μl MgCl 2 (25mM), 0.8μl dNTPs (10mM), 1μl primer YYF (10μM), 1μl primer YYR (10μM), 1μl template DNA (50ng / μl), 0.5μl Taq enzyme (2U / μl), 11.7μl ddH 2 O. The total volume is 20 μl.
[0038] The reaction program was pre-denaturation at 94°C for 4 min; 35 cycles (denaturation at 94°C for 45 s, annealing at 57°C for 45 s, extension at 72°C for 2 min); and f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com