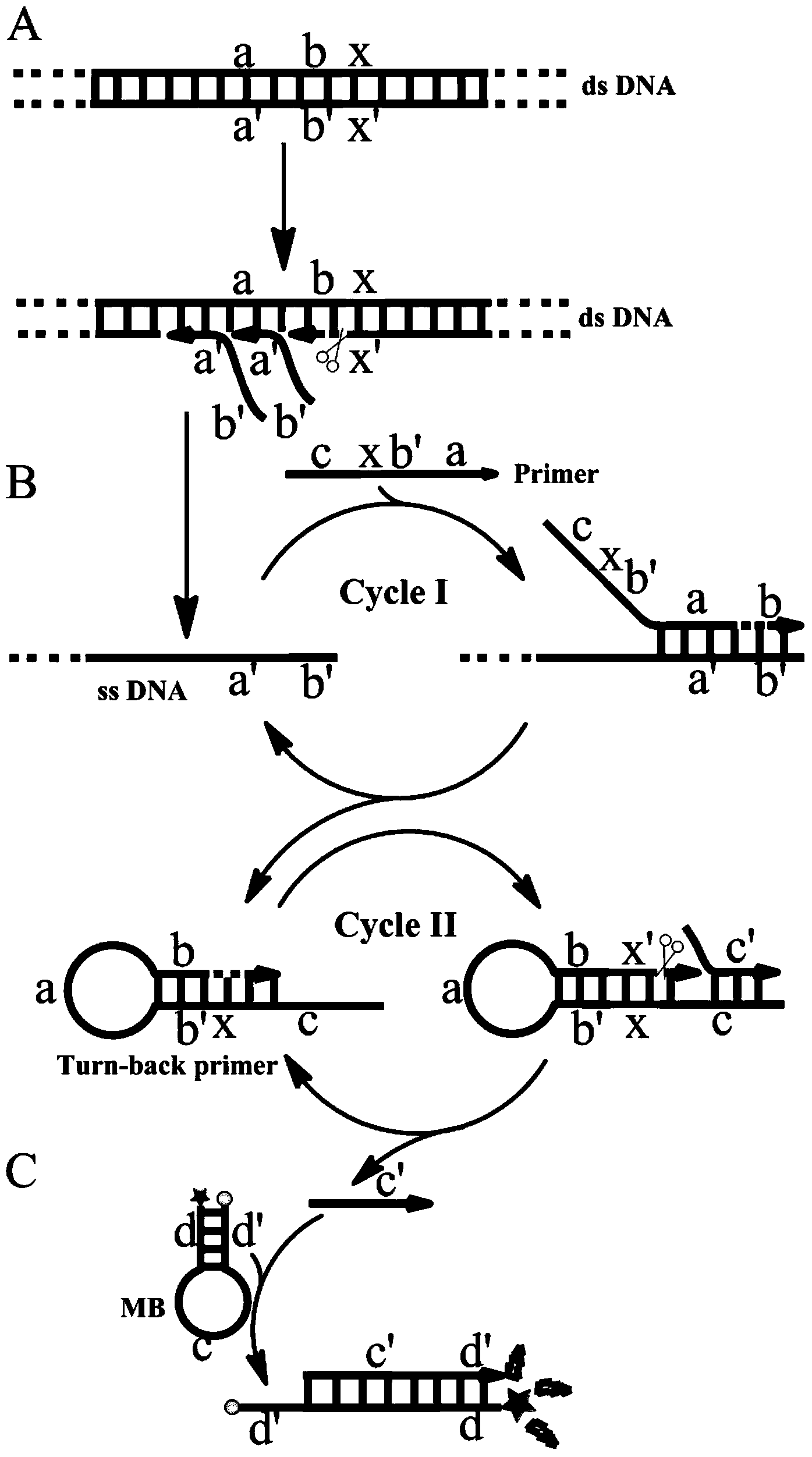
Single primer-initiated nucleic acid constant temperature amplification method
A constant-temperature amplification and single-primer technology, which is applied in biochemical equipment and methods, microbe determination/inspection, DNA/RNA fragments, etc., can solve the problems of high detection cost and complicated primer design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Embodiment 1: Verify the feasibility of the method and the correctness of its principle.
[0079] In this embodiment, PBS plasmid is used as target nucleic acid, single primer is used for constant temperature detection of amplification, and the feasibility and correctness of principle of the method are verified by fluorescent signal. Add 1 μL (5×10 -6 M) molecular beacon MB1 (sequence is 5'-FAM-CGCTTGGTAGGCTCCGGTTCCCAACGATCAGATCCTGCTACCAAGCG-DABCYL-3', which is SEQ ID NO.1), 1×NEBuffer3.1 (100mM NaCl, 50mM Tris-HCl, 10mM MgCl 2 , 100μg / mL BSA pH7.925℃), 0.05μL Bst2.0WarmStart TM DNA polymerase, 0.4 μL Nt.BstNBI nicking enzyme, 1 μL (2.5 mM) dNTPs, 0.2 μL (10 μM) primer (the sequence is 5’-GGTTCCCAACGATCAGATCCTGGTGAGACTCAACTATGCTATTTCGTTCATC-3’, which is SEQ ID NO.2), 1 μL (1×10 -12 M) The target PBS plasmid (sequence is 5'-CTATTTCGTTCATCCATAGTTGCCTGACTC-3', which is SEQ ID NO.3), and finally add water to the system to 10 μL. Using Bole CFX96 TM The real-time fluore...
Embodiment 2
[0081] Example 2: Detection of PBS plasmids using a nucleic acid constant temperature amplification detection method initiated by a single primer.
[0082] In this example, different concentrations of PBS plasmids were detected to test the sensitivity of the nucleic acid detection method. Add 1 μL (5×10 -6 M) molecular beacon MB1 (ie, SEQ ID NO.1), 1×NEBuffer3.1, 0.05μL Bst2.0WarmStart TM DNA polymerase, 0.4 μL Nt.BstNBI nickase, 1 μL (2.5 mM) dNTPs, 0.2 μL (10 μM) primer (SEQ ID NO.2), and then add 1 μL 1×10 -8 M, 1×10 -9 M, 1×10 -10 M, 1×10 -11 M, 1×10 -12 M, 1×10 -13 M, 1×10 -14 M, 1×10 -15 M, 1×10 -16 M and OM target PBS plasmids (ie, SEQ ID NO.3), add water to the system to 10 μL. Using Bole CFX96 TM The real-time fluorescent quantitative PCR instrument detects the fluorescent signal once per minute, and reacts at 55°C for 70 minutes; the results are as follows: Figure 4 As shown, the concentration gradient decreases from left to right. Figure 4 The result...
Embodiment 3
[0083] Example 3: Detection of the ability of the method to detect target nucleic acid against background interference in a complex system environment.
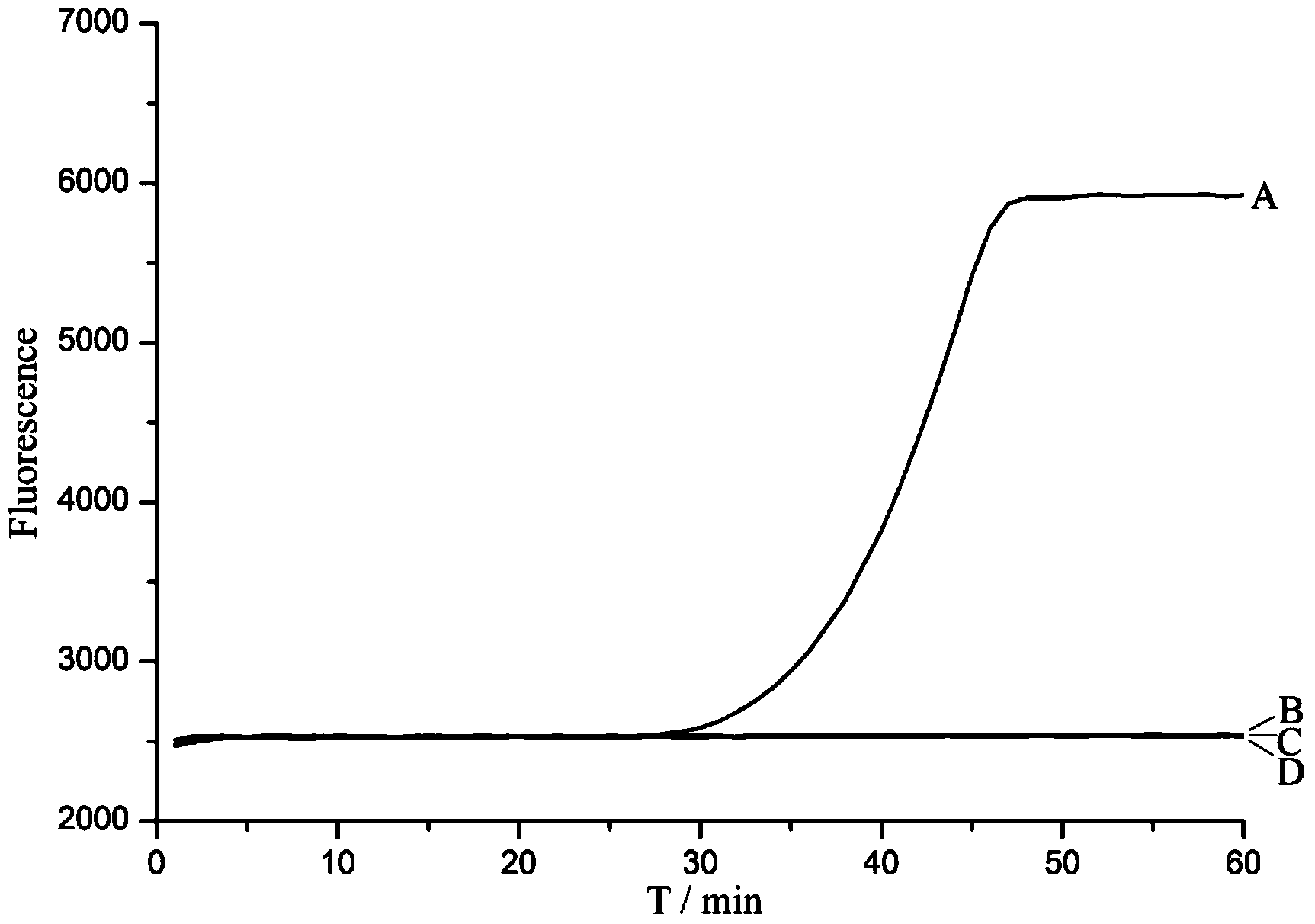
[0084] This example tests the anti-background interference ability of this scheme under the complex system containing the Escherichia coli genome. Add 1 μL (5×10 -6 M) molecular beacon MB1 (ie, SEQ ID NO.1), 1×NEBuffer3.1, 0.05μL Bst2.0WarmStart TM DNA polymerase, 0.4 μL nicking enzyme Nt.BstNBI, 1 μL (2.5 mM) dNTPs, 0.2 μL (10 μM) primer (ie, SEQ ID NO.2), 1 μL 1×10 -12 M PBS plasmid, and then add 1 μL 1×10 -10 M, 1×10 -11 M, 1×10 -12 M, 1×10 -13 M, 0M Escherichia coli genome, add water to the system to 10 μL at the end. Using Bole CFX96 TM The real-time fluorescent quantitative PCR instrument detects the fluorescent signal once every minute, and reacts at 55°C for 50 minutes. The detection result of this embodiment is as follows Figure 6 as shown, Figure 6 The middle curve results A, B, C, D, E are respectively...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com