Conjugated polymeric particle and method of making same
A polymer particle and polymer technology, applied in the field of conjugated polymer particles and its preparation, can solve the problems of high signal-to-noise ratio, low precision, low signal, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
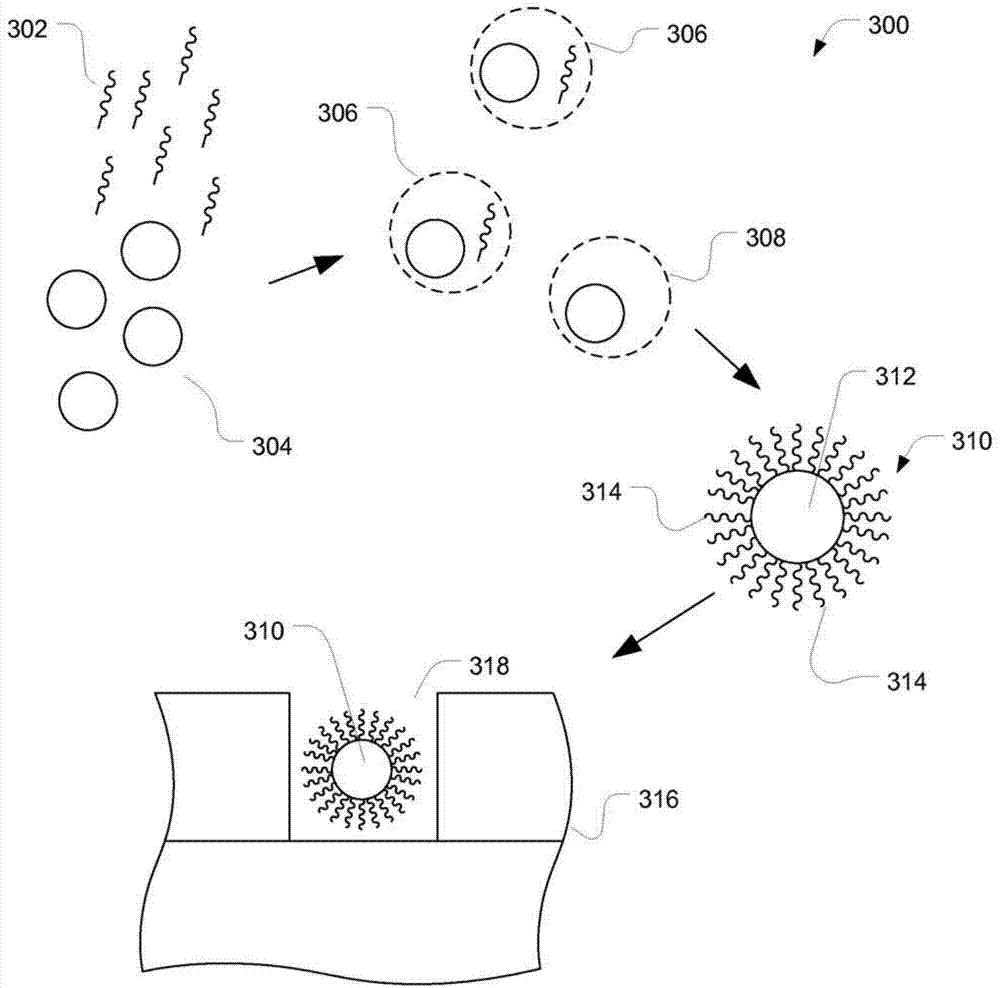
[0074] Such as Figure 4 As shown in , oligonucleotides were directly conjugated to methanesulfonyl-activated Dynal particles. Although functional groups are shown as being on the surface of the particle, functional groups may be present throughout the particle. Methanesulfonyl chloride-activated microgels were prepared via the Ugelstad process, a seeded emulsion polymerization. The particles thus formed were washed in N-methyl-2-pyrrolidone (NMP) in formulation for conjugation with ion-exchanged single-stranded DNA.
[0075] The sodium salt of the 5'-NH2-C6-30-mer oligonucleotide was dissolved in 0.1 M tetrabutylammonium acetate and injected onto a reverse phase HPLC column. Elution was performed with a 0.1 M tetrabutylammonium acetate mobile phase. Fractions containing nucleic acid were pooled, lyophilized to a dry powder, and resuspended in pure N-methyl-2-pyrrolidone (NMP).
[0076] Five million particles (5.0x10 9 ) was dispersed in 350 uL of anhydrous NMP and vortex...
Embodiment 2
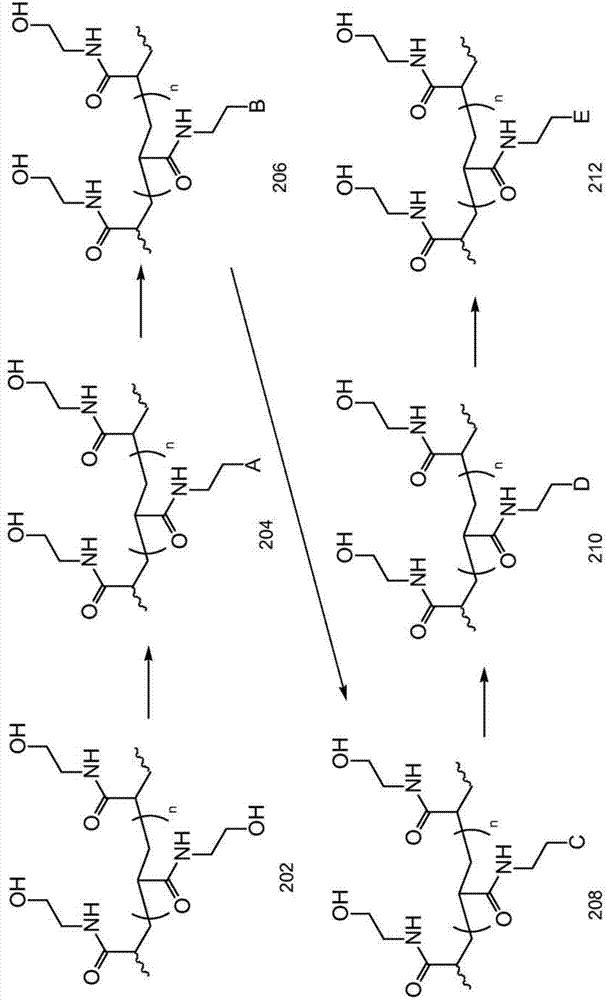
[0080] Such as Figure 5 Particles were conjugated through a series of substitutions as shown in . Although functional groups are shown as being on the surface of the particle, functional groups may be present throughout the particle.
[0081] To a methanesulfonyl chloride-activated hydrogel solution in NMP (2 billion, 1 ml) in a 2 ml centrifuge tube, a saturated solution of tetramethylammonium azide in NMP (800 uL) was added, and the reaction mixture was Stir overnight at 60°C. The reaction mixture was centrifuged at 21300 rcf for 10 minutes and the supernatant was removed. The resulting pellet was resuspended in NMP and centrifuged to remove the supernatant. This process was repeated twice.
[0082] The resulting pellet was resuspended in deionized (DI) water (1 ml). The reaction mixture was centrifuged at 21300 rcf for 10 minutes and the supernatant was removed. This process was repeated 2 more times.
[0083] 1 ml of TCEP solution (DI water, 1 M) was added to the pe...
Embodiment 3
[0087] Example 3 Activation of amino-hydrogels and conjugation of hydrogels to amine-terminal DNA probes.
[0088] Add solid disuccinyl to a solution (600 μL) of 100 million amino-hydrogels (diameter = 0.55 μm, 23 million amines / μm) in anhydrous, amine-free N-methylpyrrolidone (NMP). Amino suberate (22.1 mg, 60 μmol) followed by tributylamine (14 μL, 60 μmol). After stirring at 60°C for 1 hour, the hydrogel was isolated by centrifugation (30 minutes at 21300 rcf). The hydrogel pellet was diluted with amine-free anhydrous NMP (1 ml) and isolated by centrifugation; this washing process was repeated 2 times and the final pellet was resuspended in NMP (600 μL). The hydrogel suspension was treated with acetic anhydride (30 μL, 317 μmol) and tributylamine (30 μL, 126 μmol) and stirred at room temperature for 2 hours. The resulting hydrogel was separated by centrifugation (30 min at 21300 rcf) and the pellet was diluted with amine-free anhydrous NMP (1 ml) and separated by centrifu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com