Method for acquiring flanking sequence from genome DNA
A flanking sequence and gene sequence technology, applied in the field of obtaining flanking sequences, can solve the problems that traditional TAIL-PCR application potential cannot be brought into full play, high-efficiency TAIL-PCR is time-consuming and labor-intensive, and primer design and synthesis are expensive, so as to reduce contamination. Odds, time-saving, short-time effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1. Preparation of kit for obtaining flanking sequences from genomic DNA and method of use thereof
[0055] 1. Preparation of kits for obtaining flanking sequences from genomic DNA
[0056] The kit for obtaining flanking sequences from genomic DNA provided by the present invention comprises 15 random degenerate primers SAP, nested primers SEP and primers SNP.
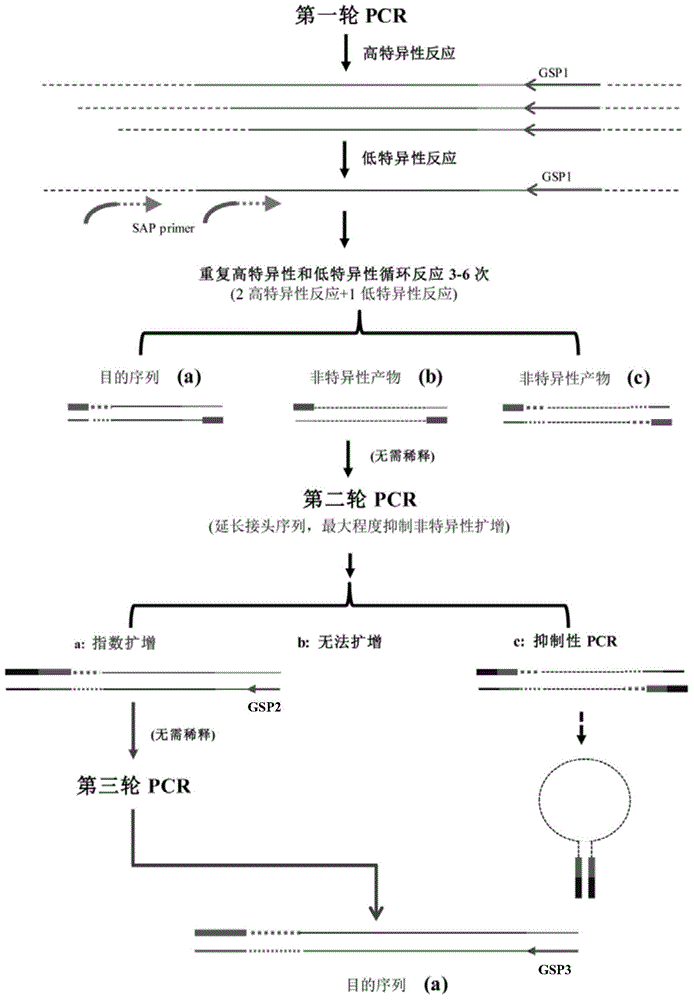
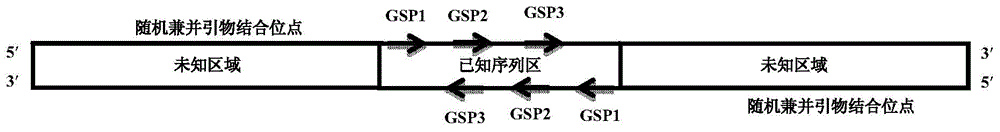
[0057] According to the principles of TAIL-PCR, suppression PCR and nested PCR, a batch of random degenerate primers (SAP1-15) with adapter sequences were designed for the first round of PCR reaction (as shown in Table 1); according to the principle of suppression PCR, Mixed nested primers (SEP) were used to extend the adapter sequence and minimize non-specific amplification; according to the principle of nested amplification, the primer SNP was designed for the third round of PCR reaction. For specific principles and procedures, see figure 1 .
[0058] Random degenerate primer SAP, nested primer SEP an...
Embodiment 2
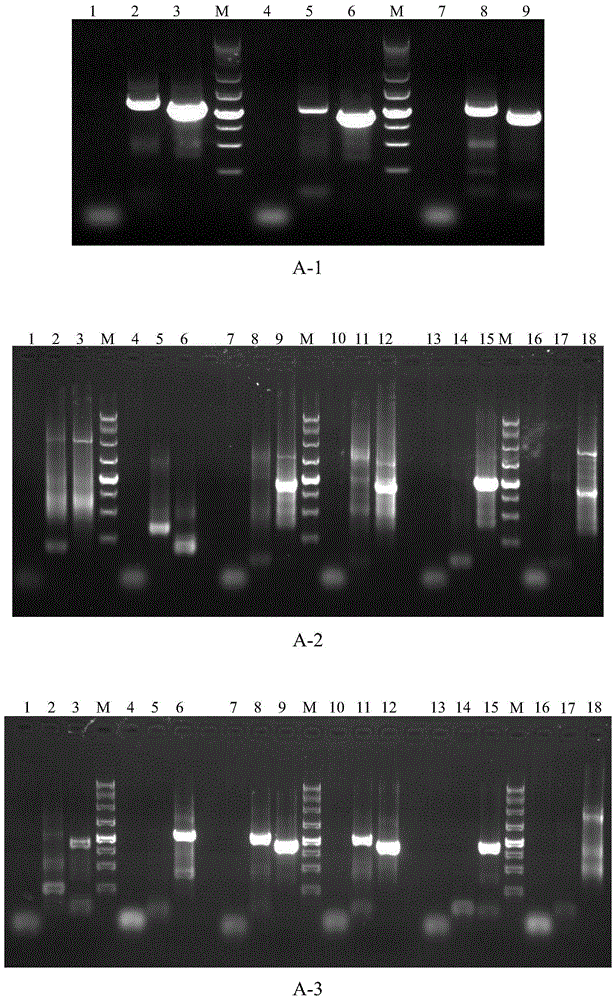
[0102] Example 2, using the kit prepared in Example 1 to obtain the 5' end sequence of LaTCTP gene from larch genomic DNA
[0103] 1. Design of LaTCTP gene-specific primers
[0104] According to the principle of specific primer design in Example 1, using primer design software primer5, three gene-specific primers 5′LaTCTP1, 5′ were designed according to the 5′ partial genome sequence (sequence 19) of larch translation regulation tumor protein LaTCTP LaTCTP2 and 5' LaTCTP3 (Table 3) were used for amplification of their 5' flanking sequences.
[0105] Table 3 LaTCTP gene-specific primers
[0106]
[0107] 2. Extraction of larch genomic DNA
[0108] Take 0.1-0.5 g of embryogenic tissue of Japanese larch (Larix leptolepis), put it in liquid nitrogen and freeze it rapidly, and use the CTAB method described in Example 1 to extract genomic DNA. Finally, 50 μL of sterilized water was used to dissolve the DNA, and the DNA concentration was determined by NanoDrop1000 to be 471.9 n...
Embodiment 3
[0127] Example 3, using the kit prepared in Example 1 to obtain the 5' end sequence of LaNFYA1 gene from larch genomic DNA
[0128] 1. Design of LaNFYA1 gene-specific primers
[0129] According to the principle of specific primer design in Example 1, using the primer design software primer5, three gene-specific primers 5'LaNFYA1-1 and 5'LaNFYA1 were designed according to the 5'UTR sequence (sequence 26) of the larch LaNFYA1 gene -2 and 5' LaNFYA1-3 (Table 4) for its 5' flanking sequence walk.
[0130] The specific primer of table 4LaNFYA1 gene
[0131]
[0132]
[0133] 2. Extraction of larch genomic DNA
[0134] With embodiment 2.
[0135] 3. The first round of PCR amplification
[0136] About 400ng of genomic DNA (about 1 μL) was used as a template, SAP1 and SAP2 (see Table 1) were selected as upstream primers, and 5′LaNFYA1-1 was used as a downstream primer, and the first round of PCR was performed.
[0137] The preparation of the first-round PCR reaction system ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com