Application of hydrogen ion transport inorganic pyrophosphatase gene expression difference on salt tolerance of tomatoes
A salt-tolerant, tomato-based technology, applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., to save time and manpower, speed up pace, and reduce workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
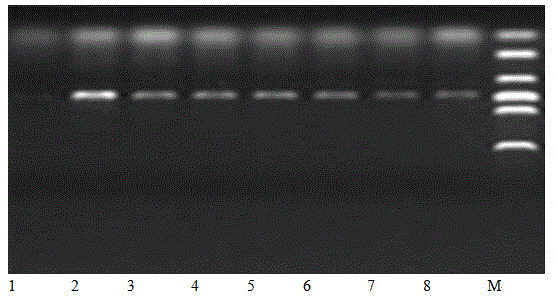
[0040] 1. The salt-tolerant tomato LA2711 and the non-salt-tolerant Tianjin local variety Jinfen 544 5-6-leaf tomato seedlings were used as materials, and the first mature leaf of the tomato plant was taken (counted from top to bottom). The detached leaves were rinsed three times with distilled water and once with deionized water. The leaves were cut into 1×2 cm rectangles with scissors, then rinsed three times with deionized water, and dried with absorbent paper. After detached leaves were stressed in 5% (w / w) sodium chloride solution for 0, 2, 4, and 6 minutes, total RNA was extracted from leaves and detected by 1% agarose gel electrophoresis.
[0041] 2. The Transcript First-Strand cDNA Synthesis SuperMix of Beijing Quanshijin Biotechnology Co., Ltd. was used for reverse transcription, and the obtained cDNA solution was used for the next step of PCR amplification.
[0042] 3. Real-time PCR
[0043] Primer design:
[0044] H + -PPase Forwoard: aatgaagagtgttggaagtgc...
Embodiment 2
[0054] Adopt specific primer to carry out the method for tomato salt tolerance determination, it is characterized in that carry out according to following steps:
[0055] (1) Take tomato seedlings with 5-6 leaves as the material, and take the first mature leaf of the tomato plant, which refers to the first mature leaf counted from top to bottom. Rinse the detached leaves three times with distilled water and once with deionized water, cut the leaves into a rectangle with a size of 1 × 2 cm with scissors, then rinse with deionized water for three times, and absorb the water with absorbent paper;
[0056] (2) Select chemically pure NaCl, 5% (w / w), and process for 2 minutes to extract total RNA from leaves;
[0057] (3) The obtained cDNA solution is used for the next step of PCR amplification;
[0058] (4) Real-time PCR
[0059] Primer design:
[0060] H + -PPase Forwoard: aatgaagagtgttggaagtgct gct
[0061] Reverse: cac gcacggtggt cttctcttca
[0062] Real-tim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com