RFLP method and kit for detecting SNP locus of MEF2C gene of cattle
A kit and scalper technology, applied in the field of molecular genetics, can solve the problems of few research reports and exploration, and achieve the effects of excellent genetic resources, low cost, and high detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] The present invention will be further described below in conjunction with the accompanying drawings and embodiments, so that the public can have an overall and sufficient understanding of the content of the invention, rather than limiting the protection scope of the present invention.
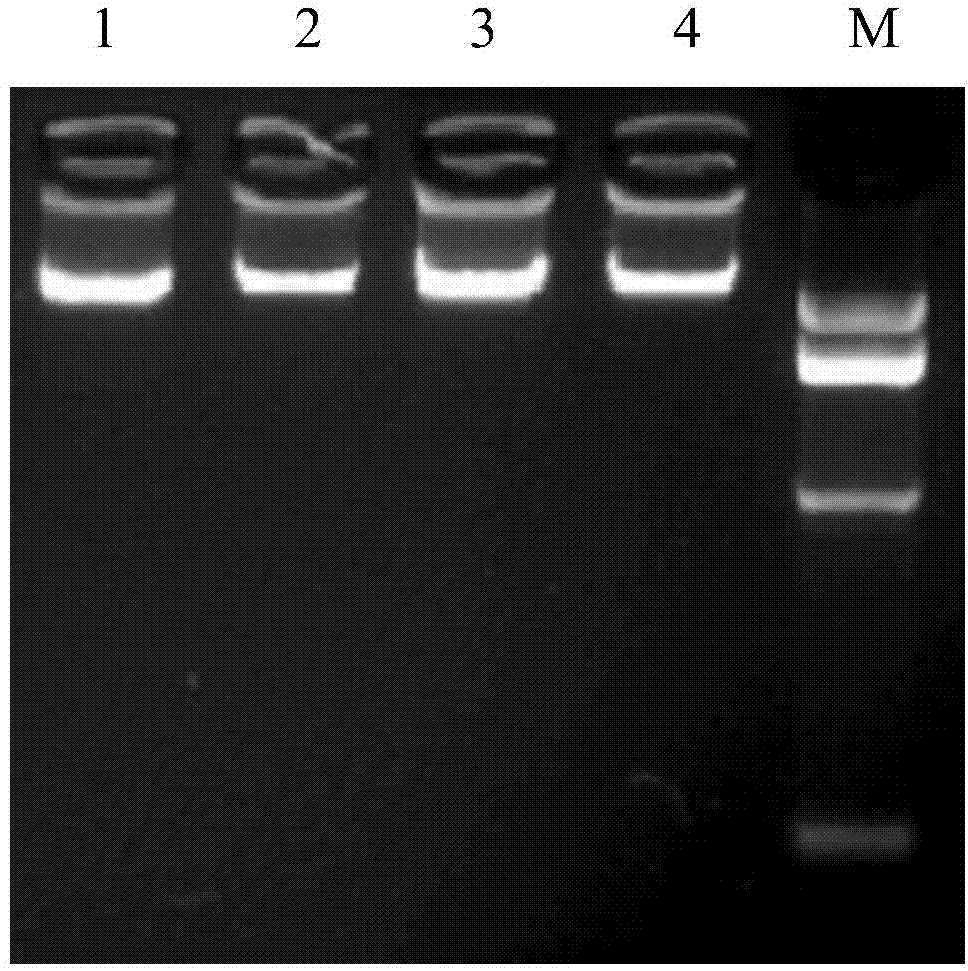
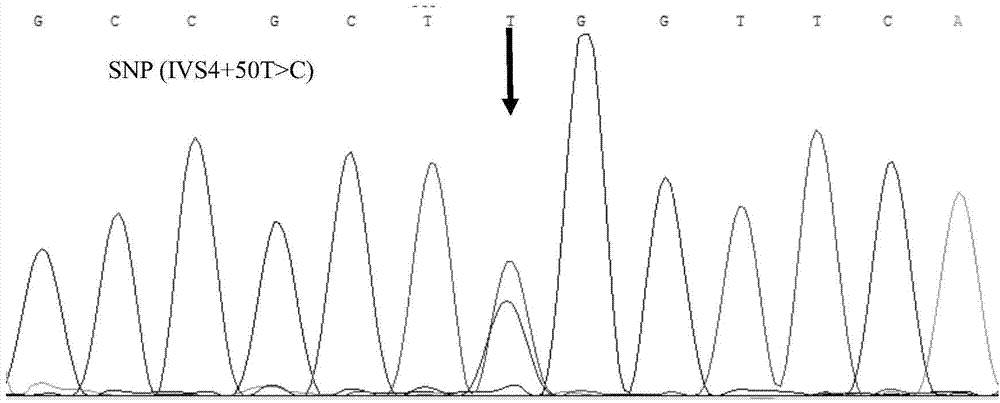
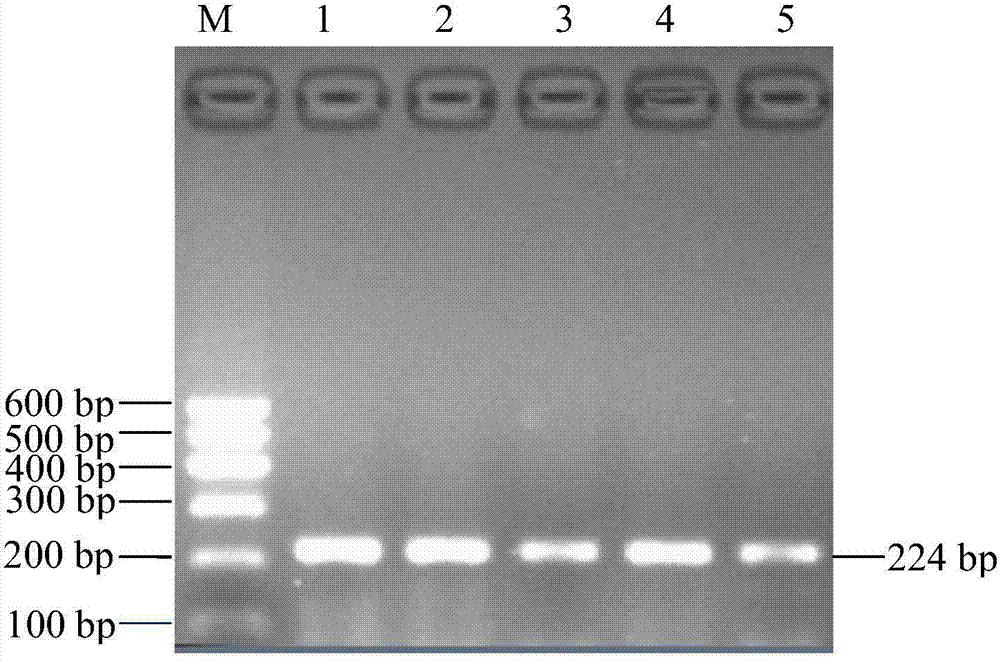
[0031] The present invention designs primers according to the MEF2C gene sequence, respectively uses the blood genome DNA of two cattle breeds as templates, performs PCR amplification, compares it with the sequence published by the authoritative database after sequencing, and finds that the 50th position of intron 4 (IVS4+50T >C) There is a single nucleotide polymorphism. For the above SNP site, the present invention detects by artificially introducing a base mismatch at the 3' end of the upstream primer through primer design. When there is no mutation at this site, the restriction endonuclease Hind Ш recognition site will be formed at the mismatched position after PCR amplification of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com