Method for porcine H11 site-specific insertion by using site specific cleavage system
A fixed-point cutting and locus technology, applied in the field of genetic engineering, can solve the problems of cumbersome steps, random insertion, and high price, and achieve the effect of simple cell screening method, high efficiency, and reduced cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
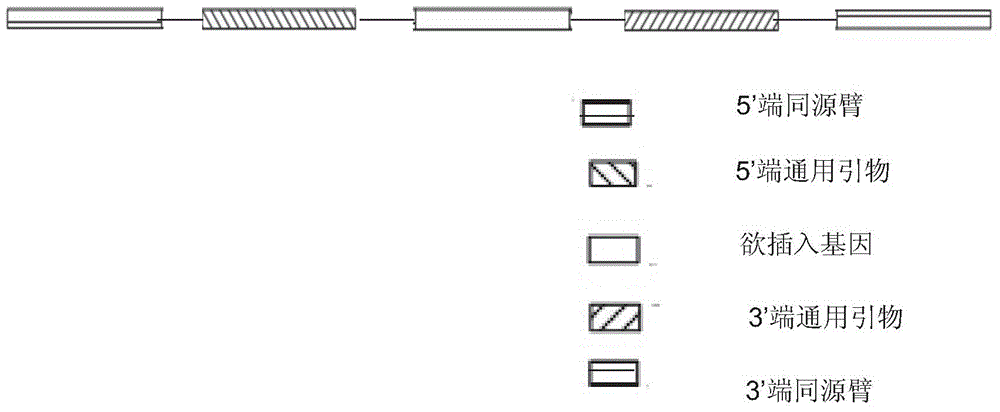
[0028] The method for inserting green fluorescent protein at the porcine H11 site by means of a site-specific cleavage system comprises the following steps:
[0029] 1. Targeting carrier construction
[0030] (1) Synthetic fragments
[0031] According to the DNA sequence of the porcine H11 site, the 3'-end homology arm (shown in sequence 3), the corresponding universal primer (shown in sequence 4) and restriction enzyme cutting sites were added at both ends: MluI (ACGCGT) and FseI ( GGCCGGCC) added, the synthetic fragment is as follows:
[0032]ACGCGTttcccgaggctGagttagttgGtccagccagtgattgagttgcgtgcggagggcttcttatcttagTTTTATAGGCTACACTGTTAACACTCAGGCTGTTTTCTACCGTTTAGTCAAAATATAGTCACCTTGCCTGCTTCACCTGTCCATCAGAGAATGGCCTCATTAATTGACTCTCTAGTATGAAGTCAAAGTAGCTTTGGTGGCCCTAAATGGACAAGTATCAAGAGACTGGGTGAATTGAGGAGCTTGAGACTGTCACCTCAGATCGAAAAGACTGAAAAATCACCTCAGATCAAAAAGACTGAAAAATCTTCAGTCTGGAAAGGGGACTCAAAACCATAATTAGAGTATTCTGGTAGAATCCTTTTCTCCACTGTTATTCATACAGTTAAGGTGAATAACTAAAAGTAATTGTGAGCTGAGGAGTAAG...
Embodiment 2
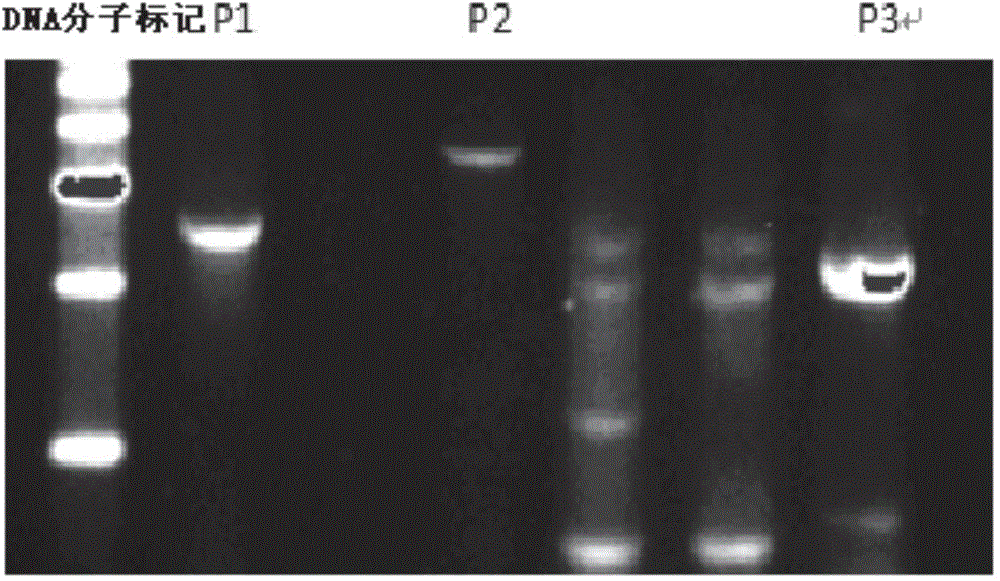
[0055] Use the same method as in Example 1 to construct a targeting vector that does not contain the gene sequence of the screening element DTA, co-transfect the PEF cells with the vector and the CRISPR / Cas9 system (the transfection system and method are the same as before), and culture at 37°C for 3 days later, filter by:
[0056] The screening method is the same as that with DTA, see Example 1 for specific operations. This method can also screen out positive clones, and a total of 192 clones were picked out, 104 of which were positive clones, with an efficiency of 54%. This experiment proved that positive clones could also be obtained without DTA.
[0057] Use the same method as in Example 1 to construct a targeting vector that does not contain the gene sequences of the screening elements DTA and Neo, and co-transfect PEF cells with the vector and the CRISPR / Cas9 system. Refer to Example 1 for the transfection system and specific methods. The DTA-free and Neo gene screening...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com