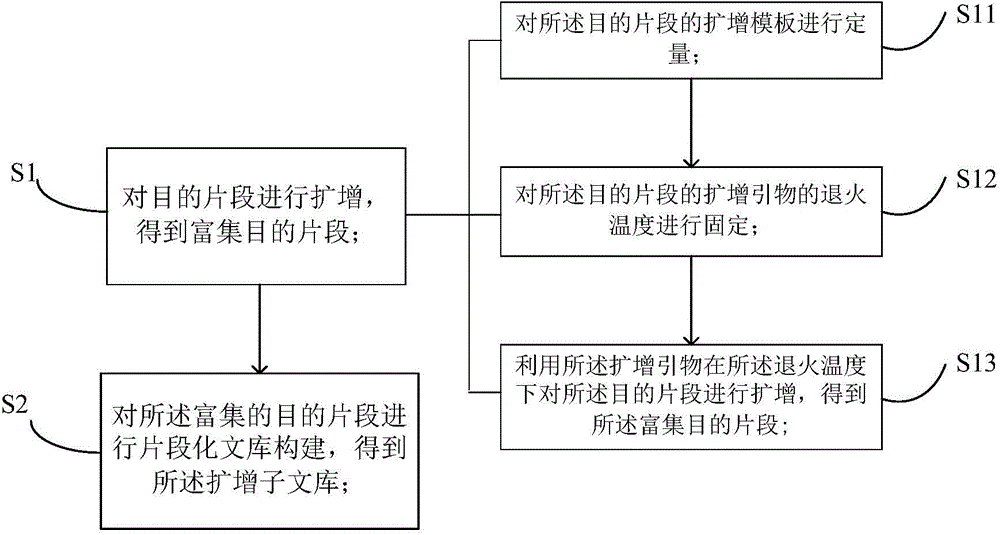
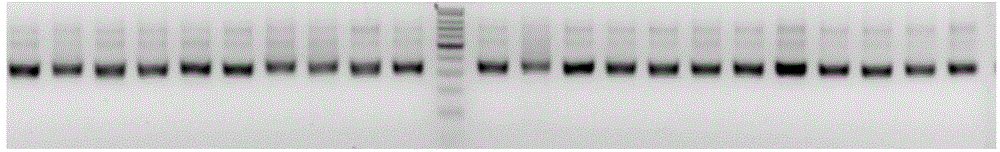
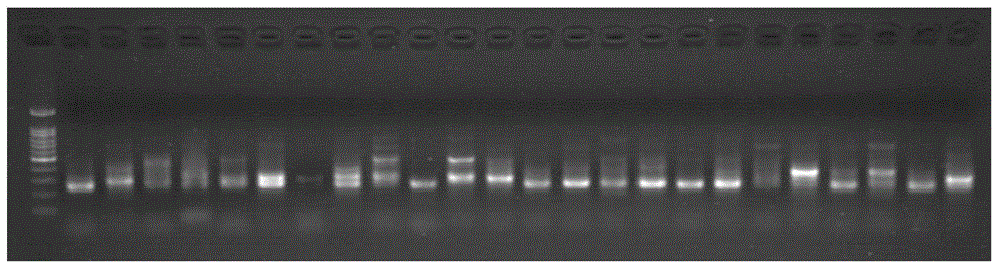
Method for constructing amplicon library
A technology of amplicon library and construction method, which is applied in the direction of chemical library, biochemical equipment and method, microorganism measurement/inspection, etc. It can solve the problems of many non-specific bands and low qualification rate, and achieve non-specific bands Less, improve the pass rate, reduce the effect of volatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1: A kind of method of 16SV4 amplicon library construction, concrete steps are as follows:
[0041] (1) Collect environmental microbial samples and extract genomic DNA from the samples. For example, using the Soil Genomic DNA Extraction Kit (DP336, Tiangen) to extract microbial DNA from soil samples.
[0042] (2) Use agarose gel electrophoresis to detect the integrity, concentration and contamination of genomic DNA. If there is a large amount of RNA in the sample, it needs to be digested with RNase. If there are many impurities in the sample (such as humic acid, organic solvent, etc.), XP magnetic beads can be used to purify the sample, and the sample can be accurately quantified in combination with a spectrophotometer or Qubit .
[0043] (3) According to the DNA concentration of the sample, the sample is diluted to reach an appropriate concentration.
[0044] (4) Amplify the hypervariable region V4 of 16S rDNA (use primers SEQ ID NO: 1: 16SV4-F-GTGCCAGCMG...
Embodiment 2
[0052] Embodiment 2: A method for constructing an ITS1 amplicon library, the main experimental steps are as follows:
[0053] (1) Collect environmental microbial samples and extract genomic DNA from the samples. For example, using the Soil Genomic DNA Extraction Kit (DP336, Tiangen) to extract microbial DNA from soil samples.
[0054] (2) Use agarose gel electrophoresis to detect the integrity, concentration and contamination of genomic DNA. If there is a large amount of RNA in the sample, it needs to be digested with RNase; if there are many impurities in the sample (such as humic acid, organic solvent, etc.), the sample can be purified using XP magnetic beads; at the same time, the sample can be accurately quantified by combining a spectrophotometer or Qubit .
[0055] (3) According to the DNA concentration of the sample, the sample is diluted to reach an appropriate concentration.
[0056] (4) Amplify ITS1 (use primers SEQ ID NO: 3: ITS1-F-GGAAGTAAAAGTCGTAACAAGG; SEQ ID ...
Embodiment 3
[0064] Embodiment 3: A method applied to the construction of ITS2 amplicon library, the main experimental steps are as follows:
[0065](1) Collect environmental microbial samples and extract genomic DNA from the samples. For example, using the Soil Genomic DNA Extraction Kit (DP336, Tiangen) to extract microbial DNA from soil samples.
[0066] (2) Use agarose gel electrophoresis to detect the integrity, concentration and contamination of genomic DNA. If there is a large amount of RNA in the sample, it needs to be digested with RNase; if there are many impurities in the sample (such as humic acid, organic solvent, etc.), the sample can be purified with XP magnetic beads. At the same time, it can be combined with a spectrophotometer or Qubit to accurately quantify the sample.
[0067] (3) According to the DNA concentration of the sample, the sample is diluted to reach an appropriate concentration.
[0068] (4) For ITS2 (use primers SEQ ID NO:5: ITS2-F-GCATCGCATAAGAACGCAGC; S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com