Method for improving PCR amplification sensitivity by using integrated host factor
A technology that integrates host factors and sensitivity, applied in the field of nucleic acid molecule amplification technology, can solve the problem that the effect of PCR amplification sensitivity is not very significant, and achieve the effect of improving sensitivity and PCR amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
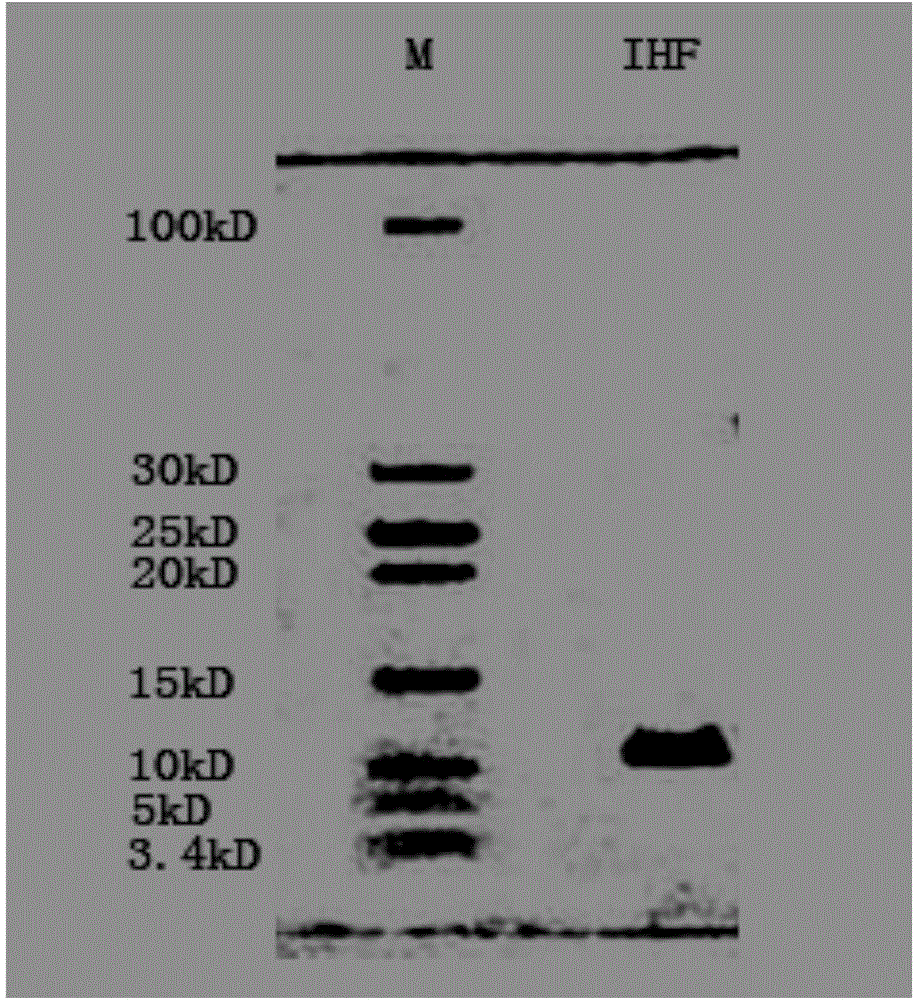
[0022] Embodiment 1: the preparation of IHF
[0023] 1. Insert IHFα or IHFβ subunits derived from Escherichia coli into PET22b expression vectors respectively, and through ligation transformation and colony PCR, enzyme digestion identification and sequencing verification, the recombinant expression vectors PET22-IHFα or pET22-IHFβ can be obtained;
[0024] 2. Transform Escherichia coli BL21 with the constructed recombinant vector pET22-IHFα or pET22-IHFβ, take the positive strain containing the recombinant plasmid and add it to LB liquid medium for overnight cultivation, and then inoculate the bacterial liquid into 50ml of LB culture medium at a ratio of 1% cultured in a shaker at 37°C until the OD value is 0.6-0.8; add IPTG, put the bacterial liquid in a shaker at 37°C to induce expression for 3 hours, and the shaker speed is 160rpm;
[0025] 3. Centrifuge the bacterial solution after the induced expression to obtain the bacterial cells, lyse and release the protein. Protein...
Embodiment 2
[0046] Embodiment 2: Optimization of PCR reaction
[0047] Use pUC19-IHF as template for PCR amplification.
[0048] Primer 1 (as shown in SEQ ID NO.2 in the sequence listing):
[0049] 5'-TATGACCAAGTCAGAATTGATA-3';
[0050] Primer 2 (as shown in SEQ ID NO.3 in the sequence listing):
[0051] 5'-CCGTAAATATTGGCGCGATCGCGC-3'.
[0052] Prepare 20 μl of PCR reaction solution according to the following table:
[0053] Reagent
Usage amount (μl)
Final concentration
2×Phusion Master Mix
10
1×
Primer1
0.1
0.018μM
Primer2
0.1
0.017μM
template DNA
0.2
0.78ng
Sterile distilled water
9.6
total capacity
20
[0054] Carry out PCR reaction according to the following conditions: 95°C for 60s. 95°C for 30s, 55°C for 30s, 72°C for 30s, a total of 30 cycles. Finally, 72°C for 5 minutes.
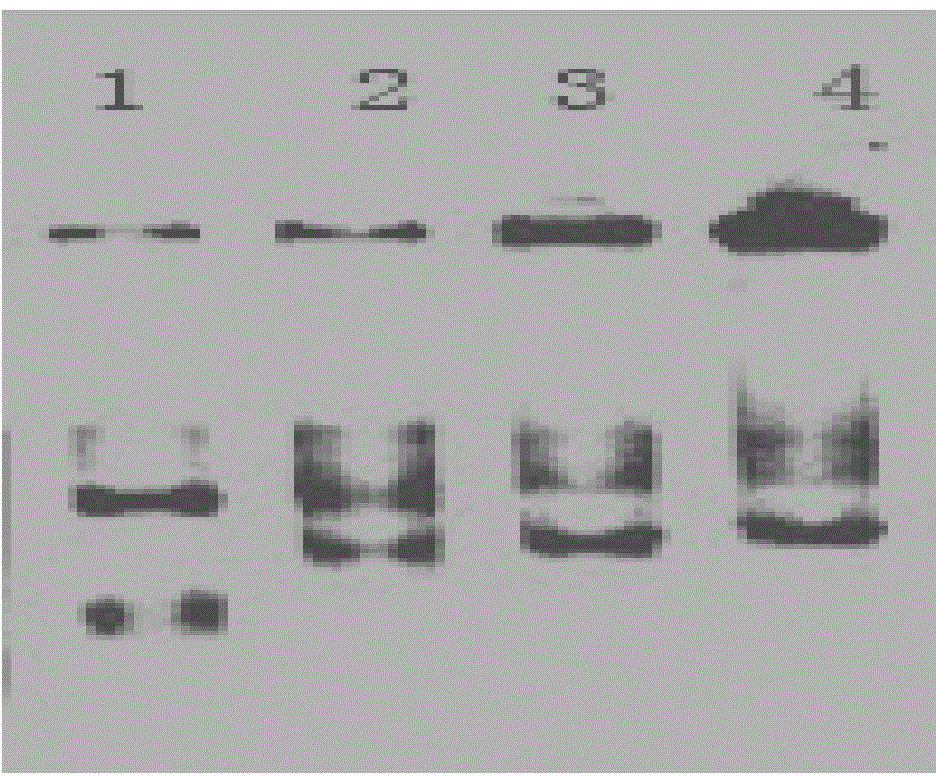
[0055] In the PCR reaction solution, different concentrations of the IHF protein obt...
Embodiment 3
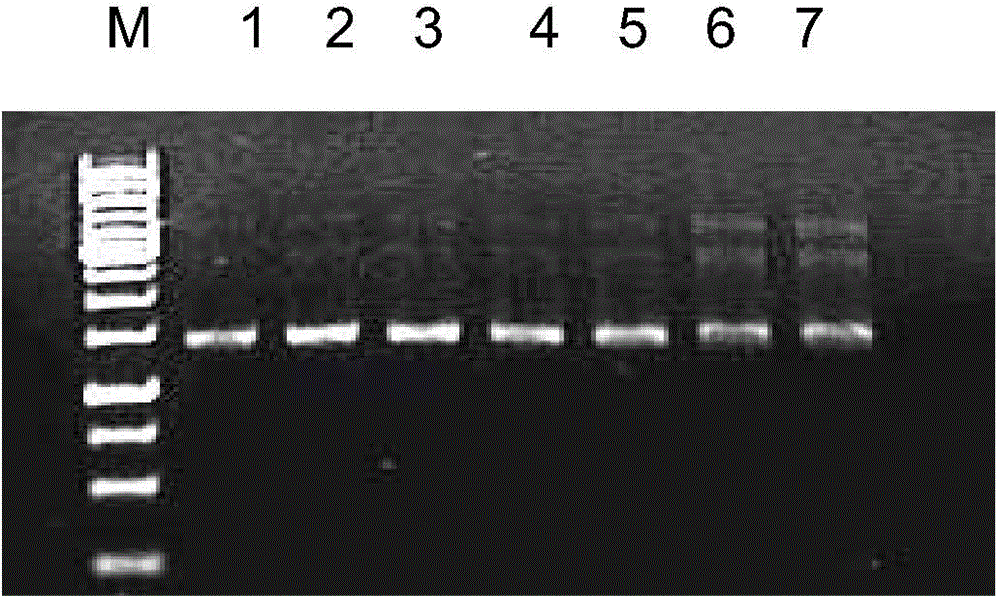
[0058] Embodiment 3: Optimization of nested PCR reaction
[0059] ①Take PFE-2-MA as a template for the first PCR
[0060] Primer 1 (shown as SEQ ID NO.4 in the sequence listing):
[0061] 5'-CTGCCGTTCATCGTTGGTGGTCGTGACGTTTCTCC-3';
[0062] Primer (shown as SEQ ID NO.5 in the sequence listing) 2:
[0063] 5'-AGAGGTAACGCAACCAGAACGACCCCAAAGAG-3'.
[0064] Prepare 20 μl of PCR reaction solution according to the following table:
[0065]
[0066] The PCR reaction was carried out according to the following conditions: 95°C for 30s, 58°C for 30s, 72°C for 50s, a total of 30 cycles. Finally, 72°C for 5 minutes.
[0067] ② Use the first PCR product diluted 100 times as the template for PCR amplification, and perform the second PCR
[0068] Primer 1 (as shown in SEQ ID NO.6 in the sequence listing):
[0069] 5'-CCTGCGCTGCTCACTGCTCTTCTGACTCTC-3';
[0070] Primer 2 (as shown in SEQ ID NO.7 in the sequence listing):
[0071] 5'-GTGGTGCATTCAGCTTCGGTCAGGATGTCA-3'.
[0072] Prepa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com