A Saccharomyces cerevisiae gene expression system and its construction and application
A Saccharomyces cerevisiae, gene expression technology, applied in the field of genetic engineering, can solve problems such as low protein expression, low plasmid copy number, and inability to meet large-scale production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
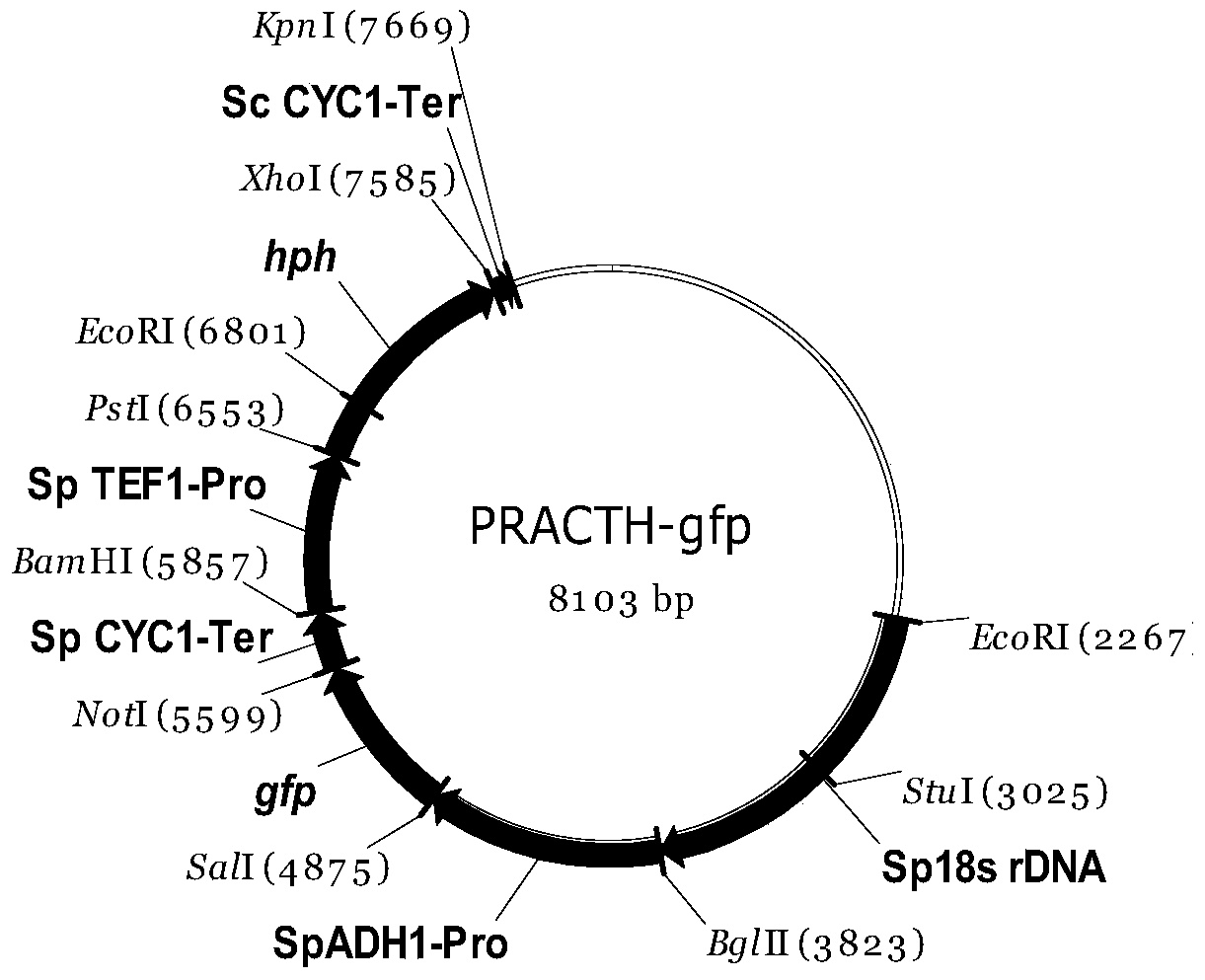
[0090] Example 1: Construction of PR series integrated expression vectors
[0091] 1. Construction of recombinant plasmid pMD-18s rDNA
[0092] (1) According to the whole genome sequence of Spathaspora passalidarum (GenBank accession NZ_AEIK00000000), two primers were designed to intercept the partial sequence of Spathaspora passalidarum 18s rDNA as the homologous recombination site. The primer sequences are as follows: the underlined part of primer P1 is the recognition site of EcoR I, and the underlined part of primer P2 is the recognition site of Bgl II, BamH I and Kpn I respectively from the 5' to 3' ends.
[0093] P1: 5'GCCG GAATTC TGCCAGTAGTCATATGCTTGTCTC3'
[0094] P2: 5'ATATTAGG GGTACC CG GGATCC GA AGATCT GTTGAAGAGCAATAAT3'
[0095] (2) Cultivate Spathaspora passalidarum overnight, collect cells, separate and extract genomic DNA.
[0096] (3) Spathaspora passalidarum genomic DNA was used as a template, and PCR amplification was performed with primers P1 and ...
Embodiment 2
[0131] Example 2: Establishment of PEG / LiAc-mediated Saccharomyces cerevisiae transformation method.
[0132] According to: Guo Zhongpeng. Metabolic Engineering Improves the Fermentation Performance of Industrial Alcoholic Yeast [D]: [Ph.D. Dissertation]. Wuxi: The method provided by Jiangnan University School of Bioengineering, 2011 prepared Saccharomyces cerevisiae ANGA1 as the host bacteria, using PEG / LiAc-mediated transformation of yeast The method is implemented as follows:
[0133] 1. Preparation of Competent State of Saccharomyces cerevisiae ANGA1
[0134] (1) Inoculate the Saccharomyces cerevisiae ANGA1 stored in the cryopreservation tube in the YPD medium, and activate the shake flask for 48 hours.
[0135] (2) Streak the activated bacterial solution on the YPD plate and store it at 4°C.
[0136] (3) Pick a single colony of Saccharomyces cerevisiae from a YPD plate, inoculate it in 20ml of YPD medium, and culture it overnight at 30°C in a 100ml shake flask.
[0137...
Embodiment 3
[0153] Example 3: Establishment of electroporation-mediated transformation of yeast Saccharomyces cerevisiae
[0154] According to: Guo Zhongpeng. Metabolic engineering to improve the fermentation performance of industrial alcohol yeast [D]: [Ph. The implementation is as follows:
[0155] 1. Preparation of Saccharomyces cerevisiae ANGA1 electroporation competent cells
[0156] (1) Inoculate the Saccharomyces cerevisiae ANGA1 stored in the cryopreservation tube into YPD medium, and activate the shake flask for 48 hours;
[0157] (2) Streak the activated bacterial solution on the YPD plate and store it at 4°C;
[0158](3) Pick a single colony of Saccharomyces cerevisiae from the YPD plate, inoculate it in 20ml YPD medium, and culture it overnight at 30°C in a 100ml shake flask;
[0159] (4) Inoculate the overnight cultured fresh bacterial solution into 50ml YPD medium, and culture it in a 250ml shake flask at 30°C and 200rpm until the OD600 of the bacterial solution reaches a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com